GPUSquad
Contents
[hide]GPUSquad
( ( (
( )\ ) )\ ) ( ( )\ )
)\ ) (()/( ( (()/( ( )\ ( )\ (()/(
(()/( /(_)) )\ /(_)))((_) )\((((_)( /(_))
/(_))_ (_)) _ ((_)(_)) ((_)_ _ ((_))\ _ )\(_))_
(_)) __|| _ \| | | |/ __| / _ \ | | | |(_)_\(_)| \
| (_ || _/| |_| |\__ \| (_) || |_| | / _ \ | |) |
\___||_| \___/ |___/ \__\_\ \___/ /_/ \_\ |___/Team Members
Progress
Assignment 1
Idea 1 - Jacobi Method for 2D Poisson Problem
This topic is based on the following pdf: https://math.berkeley.edu/~wilken/228A.F07/chr_lecture.pdf
Background:
Poisson's equation according to Wikipedia:
"In mathematics, Poisson's equation is a partial differential equation of elliptic type with broad utility in mechanical engineering and theoretical physics. It arises, for instance, to describe the potential field caused by a given charge or mass density distribution; with the potential field known, one can then calculate gravitational or electrostatic field. It is a generalization of Laplace's equation, which is also frequently seen in physics."
According to the intro in the pdf above, finite-difference and finite-element methods are the solution techniques of choice for solving elliptic PDE problems. Regardless of which type of technique you choose, you will end up with sets of linear relationships between various variables, and will need to solve for the solution u_k which satisfies all the linear relationships prescribed by the PDE.
This can be written, according to the pdf, as a matrix "Au = b, where we wish to find a solution u, given that A is a matrix capturing the differentiation operator, and b corresponds to any source or boundary terms". The Jacobi method can be used to solve this matrix, and is used in the code sample later.
Jacobi method according to Wikipedia:
"In numerical linear algebra, the Jacobi method (or Jacobi iterative method[1]) is an algorithm for determining the solutions of a diagonally dominant system of linear equations. Each diagonal element is solved for, and an approximate value is plugged in. The process is then iterated until it converges."
**************************
CODE - Based on PDF
**************************
The following code is based on the jacobi.cc and common.cc code at the bottom of the source PDF, but there are some changes for these tests. The files use .cpp extensions instead of .cc, the code in common.cc was added to jacobi.cpp so it's a single file, and code for carrying out the Jacobi iterations in the main were placed into a seperate function called dojacobi() for gprof profiling.
The std::cout line inside the output_and_error function was commented out to avoid excessive printing to the terminal. The error_and_output function was rewritten as outputImage to get rid of terminal and file I/O during calculations, and only output the final image at the end of the calculations (this would be necessary for the parallel versions, so the changes were applied to the serial code as well to make things fair).
Compilation on Linux:
g++ -std=c++0x -O2 -g -pg -o jacobi jacobi.cpp// Load standard libraries
#include <cstdio>
#include <cstdlib>
#include <iostream>
#include <fstream>
#include <cmath>
#include <chrono>
using namespace std;
// Set grid size and number of iterations
const int save_iters = 20;
const int total_iters = 5000;
const int error_every = 2;
const int m = 32, n = 1024;
const float xmin = -1, xmax = 1;
const float ymin = -1, ymax = 1;
// Compute useful constants
const float pi = 3.1415926535897932384626433832795;
const float omega = 2 / (1 + sin(2 * pi / n));
const float dx = (xmax - xmin) / (m - 1);
const float dy = (ymax - ymin) / (n - 1);
const float dxxinv = 1 / (dx*dx);
const float dyyinv = 1 / (dy*dy);
const float dcent = 1 / (2 * (dxxinv + dyyinv));
// Input function
inline float f(int i, int j) {
float x = xmin + i*dx, y = ymin + j*dy;
return abs(x)>0.5 || abs(y)>0.5 ? 0 : 1;
}
// Common output and error routine
void output_and_error(char* filename, float *a, const int sn) {
// Computes the error if sn%error every==0
if (sn%error_every == 0) {
float z, error = 0; int ij;
for (int j = 1; j<n - 1; j++) {
for (int i = 1; i<m - 1; i++) {
ij = i + m*j;
z = f(i, j) - a[ij] * (2 * dxxinv + 2 * dyyinv)
+ dxxinv*(a[ij - 1] + a[ij + 1])
+ dyyinv*(a[ij - m] + a[ij + m]);
error += z*z;
}
}
//cout << sn << " " << error*dx*dy << endl;
}
// Saves the matrix if sn<=save iters
if (sn <= save_iters) {
int i, j, ij = 0, ds = sizeof(float);
float x, y, data_float; const char *pfloat;
pfloat = (const char*)&data_float;
ofstream outfile;
static char fname[256];
sprintf(fname, "%s.%d", filename, sn);
outfile.open(fname, fstream::out
| fstream::trunc | fstream::binary);
data_float = m; outfile.write(pfloat, ds);
for (i = 0; i<m; i++) {
x = xmin + i*dx;
data_float = x; outfile.write(pfloat, ds);
}
for (j = 0; j<n; j++) {
y = ymin + j*dy;
data_float = y;
outfile.write(pfloat, ds);
for (i = 0; i<m; i++) {
data_float = a[ij++];
outfile.write(pfloat, ds);
}
}
outfile.close();
}
}
void outputImage(char* filename, float *a) {
// Computes the error if sn%error every==0
// Saves the matrix if sn<=save iters
int i, j, ij = 0, ds = sizeof(float);
float x, y, data_float; const char *pfloat;
pfloat = (const char*)&data_float;
ofstream outfile;
static char fname[256];
sprintf(fname, "%s.%d", filename, 101);
outfile.open(fname, fstream::out
| fstream::trunc | fstream::binary);
data_float = m; outfile.write(pfloat, ds);
for (i = 0; i < m; i++) {
x = xmin + i*dx;
data_float = x; outfile.write(pfloat, ds);//will need to be deferred
}
for (j = 0; j < n; j++) {
y = ymin + j*dy;
data_float = y;
outfile.write(pfloat, ds);
for (i = 0; i < m; i++) {
data_float = a[ij++];
outfile.write(pfloat, ds);
}
}
outfile.close();
}void dojacobi(int i, int j, int ij, int k, float error, float u[],
float v[], float z, float* a, float* b) {
// Set initial guess to be identically zero
for (ij = 0; ij<m*n; ij++) u[ij] = v[ij] = 0;
//output_and_error("jacobi out", u, 0);
// Carry out Jacobi iterations
for (k = 1; k <= total_iters; k++) {
// Alternately flip input and output matrices
if (k % 2 == 0) { a = u; b = v; }
else { a = v; b = u; }
// Compute Jacobi iteration
for (j = 1; j<n - 1; j++) {
for (i = 1; i<m - 1; i++) {
ij = i + m*j;
a[ij] = (f(i, j) + dxxinv*(b[ij - 1] + b[ij + 1])
+ dyyinv*(b[ij - m] + b[ij + m]))*dcent;
}
}
// Save and compute error if necessary
//output_and_error("jacobi out", a, k);
}
outputImage("jacobi out", a);
}
int main() {
int i, j, ij, k;
//float error, u[m*n], v[m*n], z;
float error, z;
float *a, *b;
float *u;
float *v;
u = new float[m*n];
v = new float[m*n];
std::chrono::steady_clock::time_point ts, te;
ts = std::chrono::steady_clock::now();
dojacobi(i, j, ij, k, error, u, v, z, a, b);
delete[] u;
delete[] v;
te = std::chrono::steady_clock::now();
std::chrono::steady_clock::duration duration = te - ts;
auto ms = std::chrono::duration_cast<std::chrono::milliseconds>(duration);
std::cout << "Serial Code Time: " << ms.count() << " ms" << std::endl;
return 0;
}Plotting Images with gnuplot
1. cd into the folder that contains the jacobi out.# files
2. run gnuplot
3. enter: set terminal png
4. enter: set output "outfilename.png"
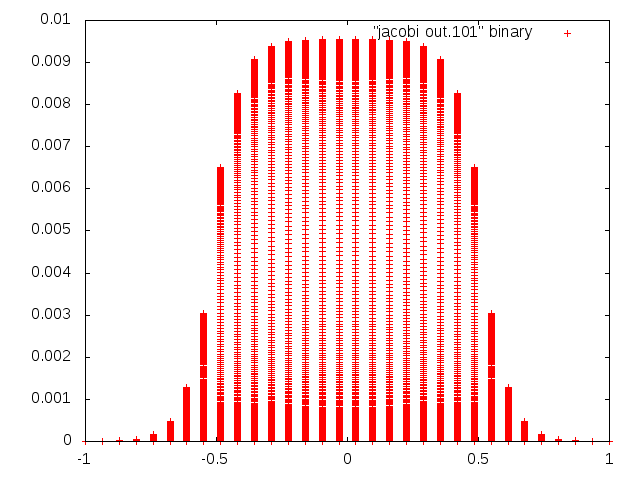
5. enter: plot "jacobi out.#" binary
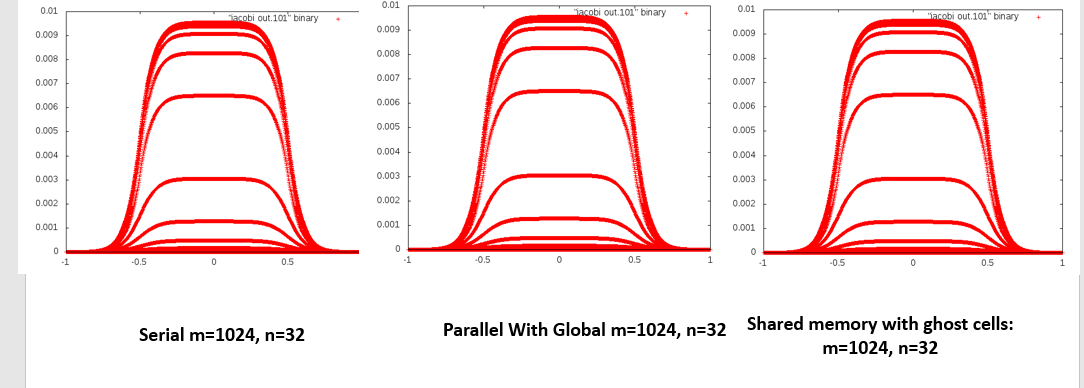
PNG image should be created in the same folder as the jacobi out.# filesThe images look something like this: m = 32, n = 1024
************************
Inputs / Performance
************************
In a 2D PDE such as the 2D Poisson problem, the matrix will have m * n gridpoints.
At m = 33, n = 33, iterations = 5000:
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls Ts/call Ts/call name
100.00 0.05 0.05 dojacobi(int, int, int, int, double, double*, double*, double, double*, double*)
0.00 0.05 0.00 5001 0.00 0.00 output_and_error(char*, double*, int)
0.00 0.05 0.00 1 0.00 0.00 _GLOBAL__sub_I__Z16output_and_errorPcPdiAt m = 165, n = 165, iterations = 5000:
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls us/call us/call name
99.15 1.16 1.16 dojacobi(int, int, int, int, double, double*, double*, double, double*, double*)
0.85 1.17 0.01 5001 2.00 2.00 output_and_error(char*, double*, int)
0.00 1.17 0.00 1 0.00 0.00 _GLOBAL__sub_I__Z16output_and_errorPcPdiAt m = 330, n = 330, iterations = 5000:
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls us/call us/call name
99.43 5.26 5.26 dojacobi(int, int, int, int, double, double*, double*, double, double*, double*)
0.57 5.29 0.03 5001 6.00 6.00 output_and_error(char*, double*, int)
0.00 5.29 0.00 1 0.00 0.00 _GLOBAL__sub_I__Z16output_and_errorPcPdi************************
The hotspot seems to be the double for-loop based on m and n in the Jacobi iterations code of the dojacobi() function. I believe these matrix calculations could be parallelized for improved performance. Note that the for-loop that the double loop is inside of is based on a constant numbers, iters, so it doesn't grow with the problem size. It would be O(iters * n^2) which is still O(n^2) not O(n^3).
Idea 2 - LZW Compression
BACKGROUND:(Paraphrased from "LZW compression" at http://whatis.techtarget.com/definition/LZW-compression) LZW compression is a compression algorithm that creates a dictionary of tokens from collections of characters. These tokens correspond to different patterns of bit values. Unique tokens are generated from the longest possible series of characters that recur in sequence any time later in a body of text. These patterns of bits that correspond to string tokens are written to an output file. Since commonly recurring sequences of words are registered to the dictionary as smaller (perhaps 12 bit) tokens, the corresponding dictionary bit code comes to represent a series of characters that would otherwise be longer than 12 bits. This is what facilitates the compression.
************************************************
CODE:
The following code is an example program that performs Compression and Decompression of text files using a LZW algorithm that encodes to 12 bit values.
This is algorithm was provided with a link as a potential project through the group projects page, but here it is again: https://codereview.stackexchange.com/questions/86543/simple-lzw-compression-algorithm
The body of code for the algorithm is as follows:
// Compile with gcc 4.7.2 or later, using the following command line:
//
// g++ -std=c++0x lzw.c -o lzw
//
//LZW algorithm implemented using fixed 12 bit codes.
#include <iostream>
#include <sstream>
#include <fstream>
#include <bitset>
#include <string>
#include <unordered_map>
#define MAX_DEF 4096
using namespace std;
string convert_int_to_bin(int number)
{
string result = bitset<12>(number).to_string();
return result;
}
void compress(string input, int size, string filename) {
unordered_map<string, int> compress_dictionary(MAX_DEF);
//Dictionary initializing with ASCII
for ( int unsigned i = 0 ; i < 256 ; i++ ){
compress_dictionary[string(1,i)] = i;
}
string current_string;
unsigned int code;
unsigned int next_code = 256;
//Output file for compressed data
ofstream outputFile;
outputFile.open(filename + ".lzw");
for(char& c: input){
current_string = current_string + c;
if ( compress_dictionary.find(current_string) ==compress_dictionary.end() ){
if (next_code <= MAX_DEF)
compress_dictionary.insert(make_pair(current_string, next_code++));
current_string.erase(current_string.size()-1);
outputFile << convert_int_to_bin(compress_dictionary[current_string]);
current_string = c;
}
}
if (current_string.size())
outputFile << convert_int_to_bin(compress_dictionary[current_string]);
outputFile.close();
}
void decompress(string input, int size, string filename) {
unordered_map<unsigned int, string> dictionary(MAX_DEF);
//Dictionary initializing with ASCII
for ( int unsigned i = 0 ; i < 256 ; i++ ){
dictionary[i] = string(1,i);
}
string previous_string;
unsigned int code;
unsigned int next_code = 256;
//Output file for decompressed data
ofstream outputFile;
outputFile.open(filename + "_uncompressed.txt");
int i =0;
while (i<size){
//Extracting 12 bits and converting binary to decimal
string subinput = input.substr(i,12);
bitset<12> binary(subinput);
code = binary.to_ullong();
i+=12;
if ( dictionary.find(code) ==dictionary.end() )
dictionary.insert(make_pair(code,(previous_string + previous_string.substr(0,1))));
outputFile<<dictionary[code];
if ( previous_string.size())
dictionary.insert(make_pair(next_code++,previous_string + dictionary[code][0]));
previous_string = dictionary[code];
}
outputFile.close();
}
string convert_char_to_string(const char *pCh, int arraySize){
string str;
if (pCh[arraySize-1] == '\0') str.append(pCh);
else for(int i=0; i<arraySize; i++) str.append(1,pCh[i]);
return str;
}
static void show_usage()
{
cerr << "Usage: \n"
<< "Specify the file that needs to be compressed or decompressed\n"
<<"lzw -c input #compress file input\n"
<<"lzw -d input #decompress file input\n"
<<"Compressed data will be found in a file with the same name but with a .lzw extension\n"
<<"Decompressed data can be found in a file with the same name and a _uncompressed.txt extension\n"
<< endl;
}
int main (int argc, char* argv[]) {
streampos size;
char * memblock;
if (argc <2)
{
show_usage();
return(1);
}
ifstream file (argv[2], ios::in|ios::binary|ios::ate);
if (file.is_open())
{
size = file.tellg();
memblock = new char[size];
file.seekg (0, ios::beg);
file.read (memblock, size);
file.close();
string input = convert_char_to_string(memblock,size);
if (string( "-c" ) == argv[1] )
compress(input,size, argv[2]);
else if (string( "-d" ) == argv[1] )
decompress(input,size, argv[2]);
else
show_usage();
}
else {
cout << "Unable to open file."<<endl;
show_usage();
}
return 0;
}
PROFILING:
The above program needs an input file to compress and decompress text in. For the purposes of testing, the Gutenberg press' "Complete Works of Shakespeare" was used as an input text file (http://www.gutenberg.org/files/100/100-0.txt) because it represents a large enough body of text to actually have perceptible run times for compression. Increases in the size of the data used are created through copying one full version of the text in the last iteration of testing and appending it to the end of the text file (so one Shakespeare's complete works becomes two back to back, two becomes four, etc).
PROFILING WITH THE ORIGINAL TEXT:
[[File:Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total time seconds seconds calls ns/call ns/call name 46.15 0.24 0.24 compress(std::string, int, std::string) 25.00 0.37 0.13 7954538 16.34 16.34 show_usage() 21.15 0.48 0.11 2091647 52.59 52.59 convert_int_to_bin(int) 7.69 0.52 0.04 2091647 19.12 35.47 std::__detail::_Map_base<std::string, std::pair<std::string const, int>, std::_Select1st<std::pair<std::string const, int> >, true, std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true> >::operator[](std::string const&) 0.00 0.52 0.00 3841 0.00 0.00 std::__detail::_Hashtable_iterator<std::pair<std::string const, int>, false, false> std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true>::_M_insert_bucket<std::pair<std::string, unsigned int> >(std::pair<std::string, unsigned int>&&, unsigned int, unsigned int) 0.00 0.52 0.00 256 0.00 0.00 std::__detail::_Hashtable_iterator<std::pair<std::string const, int>, false, false> std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true>::_M_insert_bucket<std::pair<std::string, int> >(std::pair<std::string, int>&&, unsigned int, unsigned int) 0.00 0.52 0.00 256 0.00 16.34 std::__detail::_Map_base<std::string, std::pair<std::string const, int>, std::_Select1st<std::pair<std::string const, int> >, true, std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true> >::operator[](std::string&&) 0.00 0.52 0.00 1 0.00 0.00 _GLOBAL__sub_I__Z18convert_int_to_bini]]
PROFILING WITH TWICE THE TEXT:
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total time seconds seconds calls ns/call ns/call name 38.71 0.24 0.24 compress(std::string, int, std::string) 25.81 0.40 0.16 2091647 76.49 76.49 convert_int_to_bin(int) 22.58 0.54 0.14 7954538 17.60 17.60 show_usage() 9.68 0.60 0.06 2091647 28.69 46.29 std::__detail::_Map_base<std::string, std::pair<std::string const, int>, std::_Select1st<std::pair<std::string const, int> >, true, std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true> >::operator[](std::string const&) 3.23 0.62 0.02 convert_char_to_string(char const*, int) 0.00 0.62 0.00 3841 0.00 0.00 std::__detail::_Hashtable_iterator<std::pair<std::string const, int>, false, false> std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true>::_M_insert_bucket<std::pair<std::string, unsigned int> >(std::pair<std::string, unsigned int>&&, unsigned int, unsigned int) 0.00 0.62 0.00 256 0.00 0.00 std::__detail::_Hashtable_iterator<std::pair<std::string const, int>, false, false> std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true>::_M_insert_bucket<std::pair<std::string, int> >(std::pair<std::string, int>&&, unsigned int, unsigned int) 0.00 0.62 0.00 256 0.00 17.60 std::__detail::_Map_base<std::string, std::pair<std::string const, int>, std::_Select1st<std::pair<std::string const, int> >, true, std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true> >::operator[](std::string&&) 0.00 0.62 0.00 1 0.00 0.00 _GLOBAL__sub_I__Z18convert_int_to_bini
PROFILING WITH 4X THE TEXT:
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total time seconds seconds calls us/call us/call name 40.48 1.02 1.02 compress(std::string, int, std::string) 30.16 1.78 0.76 31802927 0.02 0.02 show_usage() 22.62 2.35 0.57 8363660 0.07 0.07 convert_int_to_bin(int) 5.56 2.49 0.14 8363660 0.02 0.04 std::__detail::_Map_base<std::string, std::pair<std::string const, int>, std::_Select1st<std::pair<std::string const, int> >, true, std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true> >::operator[](std::string const&) 0.79 2.51 0.02 convert_char_to_string(char const*, int) 0.40 2.52 0.01 256 39.06 39.09 std::__detail::_Map_base<std::string, std::pair<std::string const, int>, std::_Select1st<std::pair<std::string const, int> >, true, std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true> >::operator[](std::string&&) 0.00 2.52 0.00 3841 0.00 0.00 std::__detail::_Hashtable_iterator<std::pair<std::string const, int>, false, false> std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true>::_M_insert_bucket<std::pair<std::string, unsigned int> >(std::pair<std::string, unsigned int>&&, unsigned int, unsigned int) 0.00 2.52 0.00 256 0.00 0.00 std::__detail::_Hashtable_iterator<std::pair<std::string const, int>, false, false> std::_Hashtable<std::string, std::pair<std::string const, int>, std::allocator<std::pair<std::string const, int> >, std::_Select1st<std::pair<std::string const, int> >, std::equal_to<std::string>, std::hash<std::string>, std::__detail::_Mod_range_hashing, std::__detail::_Default_ranged_hash, std::__detail::_Prime_rehash_policy, false, false, true>::_M_insert_bucket<std::pair<std::string, int> >(std::pair<std::string, int>&&, unsigned int, unsigned int) 0.00 2.52 0.00 1 0.00 0.00 _GLOBAL__sub_I__Z18convert_int_to_bini
A useful hotspot for parallelization is not immediately obvious through profiling, since the main compress() function contains the bulk of the program logic, and its show_usage(, and convert_int_to_bin are simple functions, that are called frequently in one for loop.
What really affects the runtime of the program based on data is the extent to which long matching strings can be tokenized. When this happens, larger, and larger chunks of text can be processed and compressed in an iteration. This is reflected by the fact that pasting the same blocks of text over again to increase data size does not proportionally increase run time because the same tokens work for subsequent pasted blocks.
POTENTIAL FOR PARALLELIZATION:
The compress() function performs similar operations on a collection of text, however it relies on a dictionary and an expanding string to be tokenized in a dictrionary. This could potentially be paralellized through a divide and conquer strategy where gpu blocks with shared caches share their own dictionaries and iterate over their own block of text. This, however would not be particularly useful because LZW compression relies on a globally accessible dictionary that can be continuously modified. Having multiple threads try to access the same dictionary tokens in global memory would create race conditions, and creating block-local dictionaries in shared memory would reduce the efficacy of having a large globally available dictionary to tokenize strings.
What all of this means is that LZW compression would probably be a poor candidate for parallelization due to the way that parallel memory would try to update the token dictionary.
Idea 3 - MergeSort
Based on assignment topic suggestions.
Code for logic is based on BTP500 course note by Catherine Leung - https://cathyatseneca.gitbooks.io/data-structures-and-algorithms/content/sorting/merge_sort_code.html
It has been adjust to work only with int arrays and perform operation on worst case scenario for merge sort, which is something similar to this array - [0, 2, 4, 6, 1, 3, 5, 7].
CODE
To compile on matrix - g++ -O2 -g -pg -oa1 a1.cpp
It performs merge sort on array with 100000000 which has worst case scenario positioning of elements for sorting.
#include<iostream>
int SIZE = 100000000;
/*This function merges the two halves of the array arr into tmp and then copies it back into arr*/
void merge(int arr[], int tmp[], int startA, int startB, int end) {
int aptr = startA;
int bptr = startB;
int i = startA;
while (aptr<startB && bptr <= end) {
if (arr[aptr]<arr[bptr])
tmp[i++] = arr[aptr++];
else
tmp[i++] = arr[bptr++];
}
while (aptr<startB) {
tmp[i++] = arr[aptr++];
}
while (bptr <= end) {
tmp[i++] = arr[bptr++];
}
for (i = startA; i <= end; i++) {
arr[i] = tmp[i];
}
}
//this function sorts arr from index start to end
void mSort(int* arr, int* tmp, int start, int end) {
if (start<end) {
int mid = (start + end) / 2;
mSort(arr, tmp, start, mid);
mSort(arr, tmp, mid + 1, end);
merge(arr, tmp, start, mid + 1, end);
}
}
void mergeSort(int* arr, int size) {
int* tmp = new int[size];
mSort(arr, tmp, 0, size - 1);
delete[] tmp;
}
void printArr(int* arr) {
for (int i = 0; i < SIZE; i++) {
std::cout << arr[i] << std::endl;
}
}
int main(){
int* sampleArr = new int[SIZE];
bool worstCaseFlag = false;
int j = 1;
for (int i = 0; i < SIZE; i++) {
if (worstCaseFlag == false) {
sampleArr[i] = i;
worstCaseFlag = true;
}
else {
sampleArr[i] = j;
j += 2;
worstCaseFlag = false;
}
}
mergeSort(sampleArr, SIZE);
//printArr(sampleArr);
std::cin.get();
return 0;
}
Profiling
Profiling with 1000000 elements
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls ms/call ms/call name
69.23 0.09 0.09 999999 0.00 0.00 merge(int*, int*, int, int, int)
30.77 0.13 0.04 1 40.00 130.00 mSort(int*, int*, int, int)
0.00 0.13 0.00 1 0.00 0.00 _GLOBAL__sub_I_SIZE
Profiling with 10000000 elements
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls s/call s/call name
88.89 1.04 1.04 9999999 0.00 0.00 merge(int*, int*, int, int, int)
11.11 1.17 0.13 1 0.13 1.17 mSort(int*, int*, int, int)
0.00 1.17 0.00 1 0.00 0.00 _GLOBAL__sub_I_SIZE
Profiling with 100000000 elements
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls s/call s/call name
90.40 13.85 13.85 99999999 0.00 0.00 merge(int*, int*, int, int, int)
9.60 15.32 1.47 1 1.47 15.32 mSort(int*, int*, int, int)
0.00 15.32 0.00 1 0.00 0.00 _GLOBAL__sub_I_SIZEAnalysing
The most time consuming part is merging, which can be at least partially paralleled. The Big(O) of this case is O(n).
Assignment 2
We chose to move forward with the Jacobi Method for 2D Poisson Equations code.
We parallelized the original code by placing the jacobi calculations into a kernel. For this initial parallel version, we only used 1D threading and had each thread run a for loop for the other dimension.
The iters loop launches a kernel for each iteration and we use double buffering (where we choose to launch the kernel with either d_a, d_b or d_b, d_a) since we can't simply swap pointers like in the serial code.
Double buffering is needed so that there is a static version of the the matrix before the current set of calculations are done. Since an element of the matrix is calculated based on elements on all four sides, if one of those elements updated itself while calculations were being done on another element, you would end up with the wrong answer/race conditions.
****************
CODE
****************
#include "cuda_runtime.h"
#include "device_launch_parameters.h"
#include <stdio.h>
// Load standard libraries
#include <cstdio>
#include <cstdlib>
#include <iostream>
#include <fstream>
#include <cmath>
#include <chrono>
using namespace std;__global__ void matrixKernel(float* a, float* b, float dcent, int n, int m, int xmin, int xmax,
int ymin, int ymax, float dx, float dy, float dxxinv, float dyyinv) {//MODIFY to suit algorithm
int j = blockDim.x * blockIdx.x + threadIdx.x + 1;
//above: we are using block and thread indexes to replace some of the iteration logic
if (j < n - 1) {
for (int i = 1; i < m - 1; i++) {
int ij = i + m*j;
float x = xmin + i*dx, y = ymin + j*dy;
float input = abs(x) > 0.5 || abs(y) > 0.5 ? 0 : 1;
a[ij] = (input + dxxinv*(b[ij - 1] + b[ij + 1])
+ dyyinv*(b[ij - m] + b[ij + m]))*dcent;
}
}
}
// Set grid size and number of iterations
const int save_iters = 20;
const int total_iters = 5000;
const int error_every = 2;
const int m = 32, n = 1024;
const float xmin = -1, xmax = 1;
const float ymin = -1, ymax = 1;
// Compute useful constants
const float pi = 3.1415926535897932384626433832795;
const float omega = 2 / (1 + sin(2 * pi / n));
const float dx = (xmax - xmin) / (m - 1);
const float dy = (ymax - ymin) / (n - 1);
const float dxxinv = 1 / (dx*dx);
const float dyyinv = 1 / (dy*dy);
const float dcent = 1 / (2 * (dxxinv + dyyinv));
// Input function
inline float f(int i, int j) {
float x = xmin + i*dx, y = ymin + j*dy;
return abs(x) > 0.5 || abs(y) > 0.5 ? 0 : 1;
}
// Common output and error routine
void outputImage(char* filename, float *a) {
// Computes the error if sn%error every==0
// Saves the matrix if sn<=save iters
int i, j, ij = 0, ds = sizeof(float);
float x, y, data_float; const char *pfloat;
pfloat = (const char*)&data_float;
ofstream outfile;
static char fname[256];
sprintf(fname, "%s.%d", filename, 101);
outfile.open(fname, fstream::out
| fstream::trunc | fstream::binary);
data_float = m; outfile.write(pfloat, ds);
for (i = 0; i < m; i++) {
x = xmin + i*dx;
data_float = x; outfile.write(pfloat, ds);
}
for (j = 0; j < n; j++) {
y = ymin + j*dy;
data_float = y;
outfile.write(pfloat, ds);
for (i = 0; i < m; i++) {
data_float = a[ij++];
outfile.write(pfloat, ds);
}
}
outfile.close();
}
void dojacobi() {
int i, j, ij, k;
float error, z;
float *a, *b;
float *u;
float *v;
u = new float[m*n];
v = new float[m*n];
// Set initial guess to be identically zero
for (ij = 0; ij < m*n; ij++) u[ij] = v[ij] = 0;
a = v; b = u;
float* d_a;
float* d_b;
//malloc
cudaMalloc((void**)&d_a, m*n * sizeof(float));
cudaMalloc((void**)&d_b, m*n * sizeof(float));
cudaMemcpy(d_a, a, n* m * sizeof(float), cudaMemcpyHostToDevice);
cudaMemcpy(d_b, b, n* m * sizeof(float), cudaMemcpyHostToDevice);
int nblocks = n / 1024;
dim3 dGrid(nblocks);
dim3 dBlock(1024); // Carry out Jacobi iterations
for (k = 1; k <= total_iters; k++) {
if (k % 2 == 0) {
cudaError_t error = cudaGetLastError();
matrixKernel << <dGrid, dBlock >> > (d_a, d_b, dcent, n, m, xmin, xmax, ymin, ymax, dx, dy, dxxinv, dyyinv);
cudaDeviceSynchronize();
if (cudaGetLastError()) {
std::cout << "error";
}
}
else {
cudaError_t error = cudaGetLastError();
matrixKernel << <dGrid, dBlock >> > (d_b, d_a, dcent, n, m, xmin, xmax, ymin, ymax, dx, dy, dxxinv, dyyinv);
cudaDeviceSynchronize();
if (cudaGetLastError()) {
std::cout << "error";
}
}
}
cudaMemcpy(a, d_a, n* m * sizeof(float), cudaMemcpyDeviceToHost);
outputImage("jacobi out", a);
cudaFree(d_a);
cudaFree(d_b);
delete[] u;
delete[] v;
}
int main() {
std::chrono::steady_clock::time_point ts, te;
ts = std::chrono::steady_clock::now();
dojacobi();
te = std::chrono::steady_clock::now();
std::chrono::steady_clock::duration duration = te - ts;
auto ms = std::chrono::duration_cast<std::chrono::milliseconds>(duration);
std::cout << "Parallel Code Time: " << ms.count() << " ms" << std::endl;
cudaDeviceReset();
return 0;
}****************
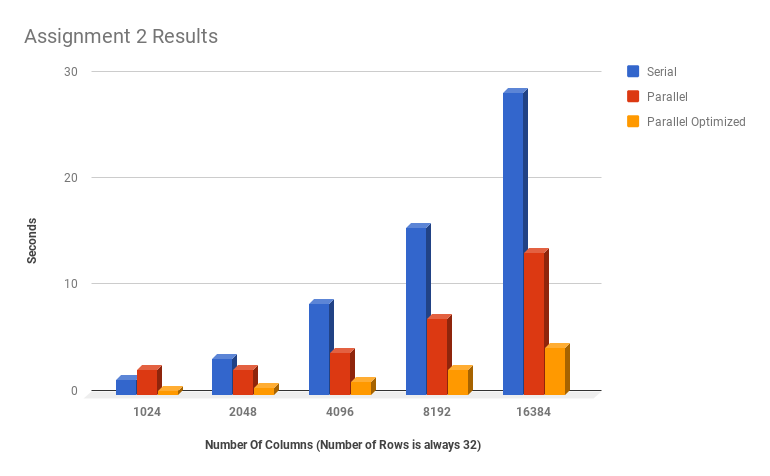
A2 TIMINGS
****************
Assignment 3
Optimization techniques attempted
- Get rid of the for loop in the kernel and use 2D threading within blocks
- Use gpu constant memory for jacobi calculation constants
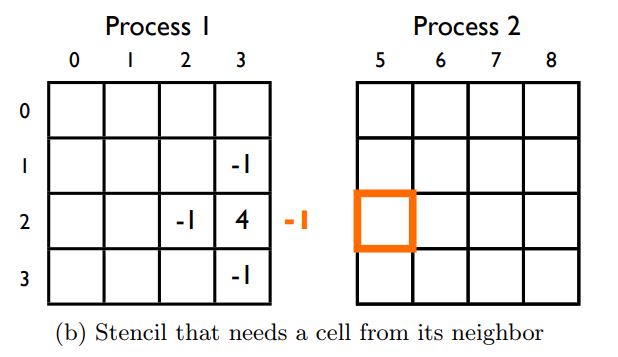
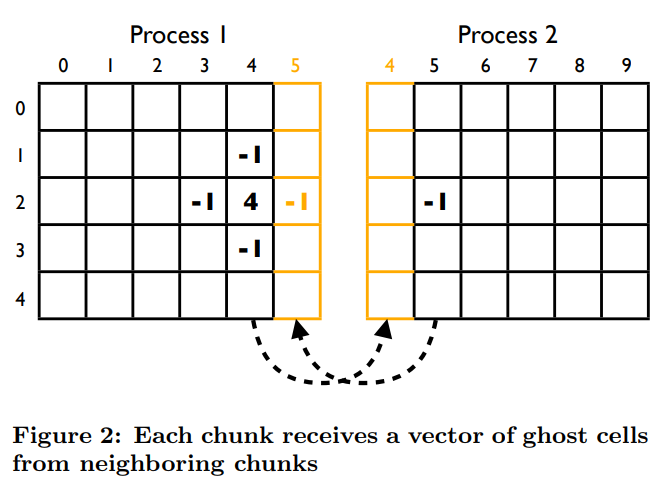
- Utilize the ghost cell pattern for shared memory within blocks
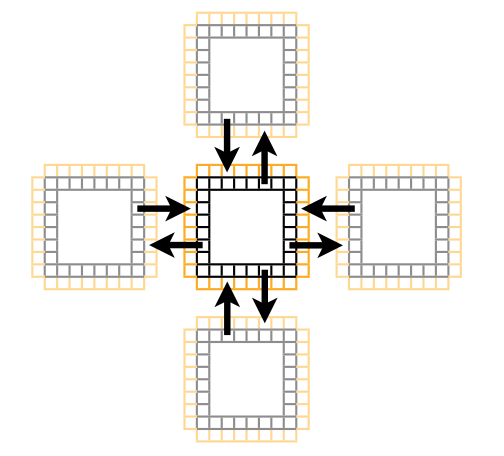
The ghost cell pattern is a technique used to allow threads in a particular block to access data that would normally be inside another block. We make a shared memory array larger than the number of threads per block in a particular dimension, then when we are at the first or last thread, can copy a neighbouring cells' data based on a global index, to our local ghost cell.
The following info and images are taken from: http://people.csail.mit.edu/fred/ghost_cell.pdf
Ghost Cell Pattern Abstract:
Many problems consist of a structured grid of points that are updated repeatedly based on the values of a fixed set of neighboring points in the same grid. To parallelize these problems we can geometrically divide the grid into chunks that are processed by different processors. One challenge with this approach is that the update of points at the periphery of a chunk requires values from neighboring chunks. These are often located in remote memory belonging to different processes. The naive implementation results in a lot of time spent on communication leaving less time for useful computation. By using the Ghost Cell Pattern communication overhead can be reduced. This results in faster time to completion.
In our code, our main jacobi calculations look like this (when using global indexing):
a[ij] = (input + dxxinv*(b[ij - 1] + b[ij + 1])
+ dyyinv*(b[ij - m] + b[ij + m]))*dcent;- b[ij - 1] is a cell's left neighbour
- b[ij + 1] is a cell's right neighbour
- b[ij - m] is a cell's top neighbour (we represent a 2D array as a 1D array, so subtract row length to go one cell up)
- b[ij + m] is a cell's bottom neighbour (add row to go one cell down)
We use a 1D grid of blocks which have 2D threads. Thus the first block needs ghost cells on the right side, the last block needs ghost cells on the left side, and all other blocks in the middle need ghost cells on both the left and right sides.
In A2 and A3 we only scaled the columns (we would have, had we not mixed up the indexing--the 2d version scales along the n dimension, while the 1d versions scale properly along the n dimension), and the other dimension always stayed at 32. This made it easy for us to keep the grid of blocks one dimensional. If we scaled the rows as well as the columns we would most likely have need to use a two dimensional grid of blocks. This would make the problem more difficult as we would then not only need ghost columns, but also ghost rows on the top and bottom of blocks. This would likely cause synchronization problems because we would need to make sure that block execution was synchronized along 2 dimensions instead of just one.
****************
CODE FOR 1D GRID WITH 2D THREAD SETUP (kernel contains shared and global memory versions)
****************
#include <cuda.h>
#include "cuda_runtime.h"
#include <device_launch_parameters.h>
#include <stdio.h>
#include <device_functions.h>
#include <cuda_runtime_api.h>
#ifndef __CUDACC__
#define __CUDACC__
#endif
// Load standard libraries
#include <cstdio>
#include <cstdlib>
#include <iostream>
#include <fstream>
#include <cmath>
#include <chrono>
using namespace std;
const int ntpb = 1024;//TOTAL number of threads per block
const int ntpbXY = 32;
__device__ __constant__ int d_m;
__device__ __constant__ int d_n;
__device__ __constant__ float d_xmin;
__device__ __constant__ float d_xmax;
__device__ __constant__ float d_ymin;
__device__ __constant__ float d_ymax;
__device__ __constant__ float d_pi;
__device__ __constant__ float d_omega;
__device__ __constant__ float d_dx;
__device__ __constant__ float d_dy;
__device__ __constant__ float d_dxxinv;
__device__ __constant__ float d_dyyinv;
__device__ __constant__ float d_dcent;
__device__ __constant__ int d_nBlocks;
__device__ __constant__ int d_ntpbXY;
void check(cudaError_t err) {
if (err != cudaSuccess)
std::cerr << "ERROR: *** " << cudaGetErrorString(err) << " ****" << std::endl;
}//every thread does a single calculation--remove iterations
//-take everything from one dimensional global, and put into 2d shared--shared has ghost b/c need for threads--
//--global is 1d indexed ,shared is 2d indexed.
//TODO: remove for loop from kernel and index entirely with blockIdx, blockDim, and threadIdx (check workshop 7 for indexing by shared memory within blocks)
__global__ void matrixKernel(float* a, float* b) {//MODIFY to suit algorithm
//int j= blockDim.x*blockIdx.x + threadIdx.x+1;//TODO: CHECK THIS DOESN'T MESS UP GLOBAL EXECUTION
int j = threadIdx.y;
int i = blockDim.x*blockIdx.x + threadIdx.x;
//int i = threadIdx.y + 1;//original code has k = 1 and <=total_iters--since k will now start at 0, set end at < total_iters
//above: we are using block and thread indexes to replace some of the iteration logic
int tj = threadIdx.y;
int ti = threadIdx.x;
int tij = threadIdx.x + ntpbXY*threadIdx.y;
int ij = i + d_m*j;
//GLOBAL MEMORY VERSION (COMMENT OUT SHARED SECTION BELOW TO RUN):==================================
// if (!(blockIdx.x == 0 && threadIdx.x == 0 || blockIdx.x == (d_nBlocks - 1) && threadIdx.x == (d_ntpbXY - 1)) && threadIdx.y != 0 && threadIdx.y != (d_ntpbXY - 1)) {
// float x = d_xmin + i*d_dx, y = d_ymin + j*d_dy;
// float input = abs(x) > 0.5 || abs(y) > 0.5 ? 0 : 1;
// a[ij] = (input + d_dxxinv*(b[ij - 1] + b[ij + 1])
// + d_dyyinv*(b[ij - d_m] + b[ij + d_m]))*d_dcent;
// }
//SHARED MEMORY SETUP (COMMENT OUT FOR GLOBAL VERSION)
__shared__ float bShared[ntpbXY+2][ntpbXY];
//NOTES ON SHARED INDEX OFFSET:
//x offset is -1, so to get the current index for the thread for global, it is bShared[ti+i][tj]
//-to get global x index -1 is bShared[ti][tj]
//-to get global x index is bShared[ti+1][tj]
//-to get global x index +1 is bShared[ti+2][tj]
//set left ghost cells
if (threadIdx.x == 0 && blockIdx.x != 0)
bShared[ti][tj] = b[ij - 1];
//set right ghost cells
if (threadIdx.x == ntpbXY - 1 && blockIdx.x != d_nBlocks - 1)
bShared[ti+2][tj] = b[ij+1];
//set ghost cell for current thread relative to global memory
bShared[ti+1][tj] = b[d_m* j + i];
__syncthreads();
//SHARED MEMORY VERSION:==========
if (!(blockIdx.x == 0 && threadIdx.x == 0 || blockIdx.x == (d_nBlocks - 1) && threadIdx.x == (d_ntpbXY - 1)) && threadIdx.y != 0 && threadIdx.y != (d_ntpbXY - 1)) {
float x = d_xmin + i*d_dx, y = d_ymin + j*d_dy;
float input = abs(x) > 0.5 || abs(y) > 0.5 ? 0 : 1;
a[d_m* j + i] = (input + d_dxxinv*(bShared[ti][tj] + bShared[ti+2][tj])//TODO: does the program logic allow for this to go out of range??
+ d_dyyinv*(bShared[ti+1][tj-1] + bShared[ti+1][tj+1]))*d_dcent;
__syncthreads();
}
}
// Set grid size and number of iterations
const int save_iters = 20;
const int total_iters = 5000;
const int error_every = 2;
//HERE:
const int m = 32, n =32 ;
const float xmin = -1, xmax = 1;
const float ymin = -1, ymax = 1;
// Compute useful constants
const float pi = 3.1415926535897932384626433832795;
const float omega = 2 / (1 + sin(2 * pi / n));
const float dx = (xmax - xmin) / (m - 1);//TODO: modify to fit kernel?
const float dy = (ymax - ymin) / (n - 1);//TODO: modify to fit kernel?
const float dxxinv = 1 / (dx*dx);
const float dyyinv = 1 / (dy*dy);
const float dcent = 1 / (2 * (dxxinv + dyyinv));
// Input function
inline float f(int i, int j) {
float x = xmin + i*dx, y = ymin + j*dy;
return abs(x) > 0.5 || abs(y) > 0.5 ? 0 : 1;
}
// Common output and error routine
void outputImage(char* filename, float *a) {
// Computes the error if sn%error every==0
// Saves the matrix if sn<=save iters
int i, j, ij = 0, ds = sizeof(float);
float x, y, data_float; const char *pfloat;
pfloat = (const char*)&data_float;
ofstream outfile;
static char fname[256];
sprintf(fname, "%s.%d", filename, 101);
outfile.open(fname, fstream::out
| fstream::trunc | fstream::binary);
data_float = m; outfile.write(pfloat, ds);
for (i = 0; i < m; i++) {
x = xmin + i*dx;
data_float = x; outfile.write(pfloat, ds);
}
for (j = 0; j < n; j++) {
y = ymin + j*dy;
data_float = y;
outfile.write(pfloat, ds);
for (i = 0; i < m; i++) {
data_float = a[ij++];
outfile.write(pfloat, ds);
}
}
outfile.close();
}
void dojacobi() {
int i, j, ij, k;
float error, z;
float *a, *b;
float *u;
float *v;
u = new float[m*n];
v = new float[m*n];
// Set initial guess to be identically zero
for (ij = 0; ij < m*n; ij++) u[ij] = v[ij] = 0;
a = v; b = u;
float* d_a;
float* d_b;
int nBlocks = ((n*m + ntpb - 1) / ntpb);
//malloc
cudaMalloc((void**)&d_a, m*n * sizeof(float));
cudaMalloc((void**)&d_b, m*n * sizeof(float));
cudaMemcpy(d_a, a, n* m * sizeof(float), cudaMemcpyHostToDevice);
cudaMemcpy(d_b, b, n* m * sizeof(float), cudaMemcpyHostToDevice);
dim3 dGrid(nBlocks, 1);//1D grid of blocks (means we don't have to ghost 1st and last rows of matrices w/i a block
dim3 dBlock(32, 32);
cudaMemcpyToSymbol(d_nBlocks, &nBlocks, sizeof(int));
cudaMemcpyToSymbol(d_ntpbXY, &ntpbXY, sizeof(int));
// Carry out Jacobi iterations
for (k = 1; k <= total_iters; k++) {
if (k % 2 == 0) {
cudaError_t error = cudaGetLastError();
//RUN KERNEL:
matrixKernel << <dGrid, dBlock >> > (d_a, d_b);
cudaDeviceSynchronize();
check(cudaGetLastError());
}
else {
cudaError_t error = cudaGetLastError();
matrixKernel << <dGrid, dBlock >> > (d_b, d_a);
cudaDeviceSynchronize();
if (cudaGetLastError()) {
std::cout << "error";
}
}
}
cudaMemcpy(a, d_a, n* m * sizeof(float), cudaMemcpyDeviceToHost);
outputImage("jacobi out", a);
cudaFree(d_a);
cudaFree(d_b);
delete[] u;
delete[] v;
}
int main() {
cudaError_t cuerr;
cuerr = cudaMemcpyToSymbol(d_m, &m, sizeof(int));
if (cudaGetLastError()) {
std::cout << cudaGetErrorString(cuerr) << std::endl;
} cudaMemcpyToSymbol(d_n, &n, sizeof(int));
cudaMemcpyToSymbol(d_xmin, &xmin, sizeof(float));
cudaMemcpyToSymbol(d_xmax, &xmax, sizeof(float));
cudaMemcpyToSymbol(d_ymin, &ymin, sizeof(float));
cudaMemcpyToSymbol(d_ymax, &ymax, sizeof(float));
cudaMemcpyToSymbol(d_pi, &pi, sizeof(float));
cudaMemcpyToSymbol(d_omega, &pi, sizeof(float));
cudaMemcpyToSymbol(d_dx, &dx, sizeof(float));
cudaMemcpyToSymbol(d_dy, &dy, sizeof(float));
cuerr = cudaMemcpyToSymbol(d_dxxinv, &dxxinv, sizeof(float));
if (cudaGetLastError()) {
std::cout << "error" << std::endl;
std::cout << cudaGetErrorString(cuerr) << std::endl;
}
cudaMemcpyToSymbol(d_dyyinv, &dyyinv, sizeof(float));
cuerr = cudaMemcpyToSymbol(d_dcent, &dcent, sizeof(float));
if (cudaGetLastError()) {
//std::cout << "error";
std::cout << cudaGetErrorString(cuerr) << std::endl;
}
std::chrono::steady_clock::time_point ts, te;
ts = std::chrono::steady_clock::now();
dojacobi();
te = std::chrono::steady_clock::now();
std::chrono::steady_clock::duration duration = te - ts;
auto ms = std::chrono::duration_cast<std::chrono::milliseconds>(duration);
std::cout << "Parallel Code Time: " << ms.count() << " ms" << std::endl;
cudaDeviceReset();
return 0;
}****************
A NOTE ON SCALABILITY:
In our attempts to make the kernel scalable with ghost cells, we scaled along one dimension. However, we were inconsistent in our scaling. The 1D kernel scaled along the n (y) dimension while the 2d kernels scaled along the m (x) dimension. Scaling along the x dimension, while allowing results to be testable between serial and 2D parallelized versions of the code, produced distributions that were strangely banded and skewed. In other words, we made the code render weird things faster:
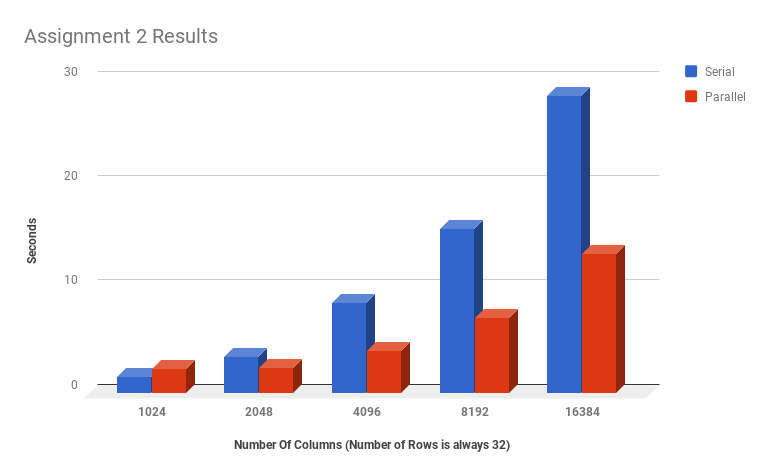
FINAL TIMINGSTHE GRAPH IMMEDIATELY BELOW IS INCORRECT: there was an error recording the 1D runtimes for assignment 2
****************
****************
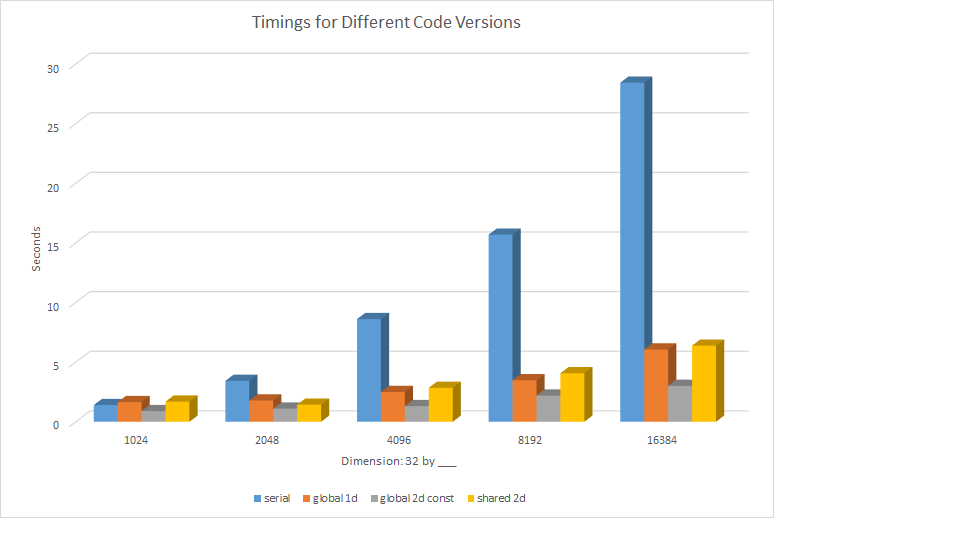
PROPER TIMINGS:
Blue = Serial (A1) Orange = Parallel (A2) Grey = Optimized Global (A3) Yellow = Optimized Shared (A3)
The above graph includes the total run times for the serial code, the 1D kernel from assignment 2, a kernel with global and constant memory with a 2D thread arrangement, and the same 2D arrangement but with shared memory utilizing ghost cells.
We found that the most efficient version of the code was the 2d version that used constant memory and did not use shared memory. Because the shared memory version of the kernel required synchronization of threads to allocate shared memory every time a kernel was run, and a kernel was run 5000 times for each version of our code, this increased overhead for memory setup actually made the execution slower than the version with global memory.
We found that the most efficient version of the code was the 2d version that used constant memory and did not use shared memory. Because the shared memory version of the kernel required synchronization of threads to allocate shared memory every time a kernel was run, and a kernel was run 5000 times for each version of our code, the if statements required to set up the ghost cells for shared memory may have created a certain amount of warp divergence, thus slowing down the runtimes of each individual kernel.
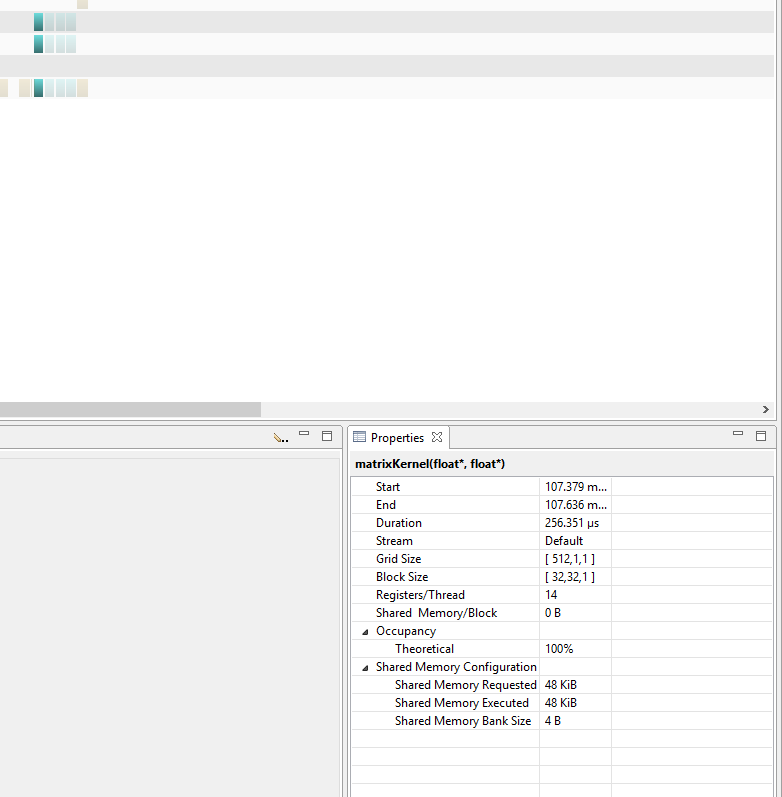
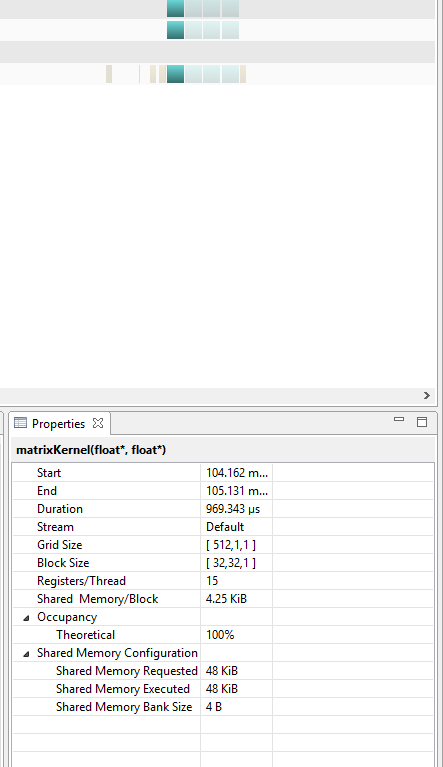
Below, are two images that show 4 consecutive kernel runs for both global and shared versions of the code. It is apparent that shared kernel runs actually take more time than the global memory versions.
TIMES FOR THE SHARED KERNEL
Note how the run times for each kernel with shared memory are significantly longer than those with global.
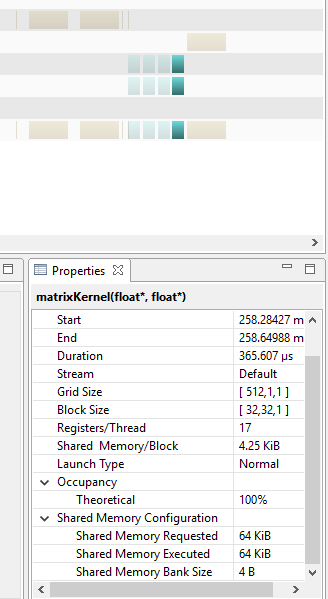
To try to determine if this issue was one of warp divergence, we tried to time a kernel with global memory that also initialized shared memory, although referenced global memory when carrying out the actual calculations:
The run of a kernel that allocated shared memory using a series of if statements, but executed instructions using global memory is shown in the figure above. While slightly longer than the run with global memory where shared memory is not initialized for ghost cells, it still takes less time to run than the version with Global memory. It is likely that Our group's attempts to employ shared memory failed because we did not adequately schedule or partition the shared memory, and the kernel was slowed as a result. The supposed occupancy of a block of shared memory was 34x32 (the dimensions of the shared memory matrix) x 4 (the size of a float) which equals 4,352 bytes per block, which is supposedly less than the maximum of about 49KB stated for a device with a 5.0 compute capability (which this series of tests on individual kernel run times was performed on). With this is mind it is still unclear as to why the shared memory performed more poorly that the global memory implementation.
Unfortunately our group's inability to effectively use profiling tools has left this discrepancy as a mystery.
In conclusion, while it may be possible to parallelize the algorithm we chose well, the effort to do so would involve ensuring that shared memory is properly synchronized in two block dimensions (2 dimensions of ghost cells rather than the 1 we implemented), and to ensure that shared memory is allocated appropriately such that maximum occupancy is established within the GPU. Unfortunately, our attempts fell short, and while implementing constant memory seemed to speed up the kernel a bit, our solution was not fully scalable in both dimensions, and shared memory was not implemented in a way that improved kernel efficiency.