Difference between revisions of "BLAStoise"
(→Assignment 1) |
(→Assignment 3) |
||
| (27 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
== Team Members == | == Team Members == | ||
# [mailto:mmbabol@senecacollege.ca?subject=gpu610 Matt Babol], Sudoku | # [mailto:mmbabol@senecacollege.ca?subject=gpu610 Matt Babol], Sudoku | ||
| − | # [mailto:jdesmond1@senecacollege.ca?subject=gpu610 Jonathan Desmond], | + | # [mailto:jdesmond1@senecacollege.ca?subject=gpu610 Jonathan Desmond], DNA Edit Distance and Alignment Tool |
| − | # [mailto:tsjiang1@senecacollege.ca?subject=gpu610 Sallie Jiang], | + | # [mailto:tsjiang1@senecacollege.ca?subject=gpu610 Sallie Jiang], Oil Painting |
[mailto:fardad.soleimanloo@senecacollege.ca,chris.szalwinski@senecacollege.ca?subject=gpu610 Email All] | [mailto:fardad.soleimanloo@senecacollege.ca,chris.szalwinski@senecacollege.ca?subject=gpu610 Email All] | ||
_ | _ | ||
| Line 46: | Line 46: | ||
=== Assignment 1 === | === Assignment 1 === | ||
| − | + | <h4>Sudoku Solver by Matt B.</h4> | |
Sudoku solver is a program that solves a sudoku puzzle. The user can input an existing file and have the program solve this, or can manually enter values to be solved. The sudoku puzzle is 9x9 in size. The data needs to be in a specific format for the program to work. There are 9 rows of values, with each cell/element in the row needing to be separated by a space. A value of 0 tells the program to solve this value. | Sudoku solver is a program that solves a sudoku puzzle. The user can input an existing file and have the program solve this, or can manually enter values to be solved. The sudoku puzzle is 9x9 in size. The data needs to be in a specific format for the program to work. There are 9 rows of values, with each cell/element in the row needing to be separated by a space. A value of 0 tells the program to solve this value. | ||
| Line 53: | Line 53: | ||
| − | + | <h5>Easy puzzle</h5> | |
| + | |||
| + | <h6>Running program</h6> | ||
To compile the program, open the terminal and go to the projects directory | To compile the program, open the terminal and go to the projects directory | ||
| Line 89: | Line 91: | ||
9 2 5 6 8 7 3 4 1 | 9 2 5 6 8 7 3 4 1 | ||
| − | + | <h6>Test Case</h6> | |
To profile the program, run this command. | To profile the program, run this command. | ||
| Line 175: | Line 177: | ||
| − | + | <h6>Analysis</h6> | |
The analysis reveals that the program took no time in solving this puzzle. However, the function checkRow and checkColumn were called the most. These two functions are used for checking whether the row and columns are correct. For a deeper analysis, a harder puzzle must be used. | The analysis reveals that the program took no time in solving this puzzle. However, the function checkRow and checkColumn were called the most. These two functions are used for checking whether the row and columns are correct. For a deeper analysis, a harder puzzle must be used. | ||
| − | + | <h5>Hard puzzle</h5> | |
| + | |||
| + | <h6>Running program</h6> | ||
For the hard puzzle, below is the input file as well as the result | For the hard puzzle, below is the input file as well as the result | ||
| Line 203: | Line 207: | ||
4 7 2 3 1 9 5 6 8 | 4 7 2 3 1 9 5 6 8 | ||
8 6 3 7 4 5 2 1 9 | 8 6 3 7 4 5 2 1 9 | ||
| + | |||
| + | <h6>Test Cases</h6> | ||
The profiling results are | The profiling results are | ||
| Line 293: | Line 299: | ||
[6] checkSquare(int, int, int) [20] __static_initialization_and_destruction_0(int, int) [3] placeNum(int, int) | [6] checkSquare(int, int, int) [20] __static_initialization_and_destruction_0(int, int) [3] placeNum(int, int) | ||
| − | + | <h6>Analysis</h6> | |
With a harder puzzle, the time for the program to solve the puzzle increased significantly. The total time to complete this puzzle was 17.13 seconds. The program spends almost half of its running time checking if the row is correct, and another 18% of its time checking whether the column is correct. This program contains thousands of calls to check the row and column values, this is why the program would be excellent project for parallelizing. | With a harder puzzle, the time for the program to solve the puzzle increased significantly. The total time to complete this puzzle was 17.13 seconds. The program spends almost half of its running time checking if the row is correct, and another 18% of its time checking whether the column is correct. This program contains thousands of calls to check the row and column values, this is why the program would be excellent project for parallelizing. | ||
| Line 300: | Line 306: | ||
---- | ---- | ||
| − | + | <h4>Oil Painting By Sallie J.</h4> | |
This program converts a regular image into a stylized oil painting, it uses OpenCV. The painting algorithm depends on the brush size and colour intensity. The program takes three command line arguments: int for brush size, int for intensity and file name of an image. Upon finishing, the program produces the original image along with the oil paint version and the total time required in seconds. | This program converts a regular image into a stylized oil painting, it uses OpenCV. The painting algorithm depends on the brush size and colour intensity. The program takes three command line arguments: int for brush size, int for intensity and file name of an image. Upon finishing, the program produces the original image along with the oil paint version and the total time required in seconds. | ||
| Line 308: | Line 314: | ||
However there have been some changes to make testing and profiling slightly easier. (Mainly changes are putting the for-loop logic into a function outside of the main and modifying for command line arguments instead of hard coding values.) | However there have been some changes to make testing and profiling slightly easier. (Mainly changes are putting the for-loop logic into a function outside of the main and modifying for command line arguments instead of hard coding values.) | ||
| − | + | <h5>Running the program</h5> | |
To compile the program on Linux you must download the OpenCV library and then create a makefile that will create the executable OilPaint.exe. To compile the program in visual studio you will need to set project properties for OpenCV through setting the C/C++ Additional Include Directories, Linker Additional Library Directories and Input Additional Dependencies (opencv_world320d.lib). | To compile the program on Linux you must download the OpenCV library and then create a makefile that will create the executable OilPaint.exe. To compile the program in visual studio you will need to set project properties for OpenCV through setting the C/C++ Additional Include Directories, Linker Additional Library Directories and Input Additional Dependencies (opencv_world320d.lib). | ||
| + | [[Media: OilPaint.zip]] | ||
| + | This zip file contains a make file and the cpp file. To compile on Linux alter the makefile. | ||
Run the executable with the arguments 5 (brush size), 20 (colour intensity), filename.format (including file format) | Run the executable with the arguments 5 (brush size), 20 (colour intensity), filename.format (including file format) | ||
| − | + | $ cmake -DCMAKE_CXX_FLAGS=-pg =DCMAKE_EXE_LINKER_FLAGS=-pg -DCMAKE_SHARED_LINKER_FLAGS=-pg -pg . | |
| + | $ make | ||
| + | $ ./OilPaint 5 20 filename.fileformat | ||
| − | + | <h6>Output:</h6> | |
| − | gprof Output: | + | [[File:t2.jpg|550px]][[File:OilVersion-t2.jpg|550px]] |
| + | |||
| + | <h6>gprof Output:</h6> | ||
Flat profile: | Flat profile: | ||
Each sample counts as 0.01 seconds. | Each sample counts as 0.01 seconds. | ||
| Line 329: | Line 341: | ||
... (there are a lot of other calls to the library that did not significantly affect the profiling) | ... (there are a lot of other calls to the library that did not significantly affect the profiling) | ||
| − | + | <h6>Visual Studio Performance profiler Output:</h6> | |
[[File:VSPROFILE.png]] | [[File:VSPROFILE.png]] | ||
The time required for the program depends largely on the file size being converted. Around 5 seconds for a 50KB image and 100 seconds for a 1MB image. It depends on the brush size and intensity levels as well. | The time required for the program depends largely on the file size being converted. Around 5 seconds for a 50KB image and 100 seconds for a 1MB image. It depends on the brush size and intensity levels as well. | ||
| − | + | <h5>Analysis</h5> | |
The profiling revealed that 80-99% of the processing time is spent in the paint function where the for-loop logic is located. Within that 99% the program spends roughly 2/3 of its time reading accessing data through the "at" function of the OpenCV Mat class (n-dimensional dense array class). The other 1/3 is spent on direct access through OpenCV’s Vec class (short numerical vectors). The for-loop is structured divides the picture up based on brush size. Then it finds the colour for each pixel in that section. Finally, it then averages the intensity to produce the final colour of that group of pixels. This is what makes this program ideal for parallelizing, because each iteration of this for-loop is calculating the final colours for each pixel. (SIMD type of process, the single instruction is to find the final colour and the multiple data is the pixels.) | The profiling revealed that 80-99% of the processing time is spent in the paint function where the for-loop logic is located. Within that 99% the program spends roughly 2/3 of its time reading accessing data through the "at" function of the OpenCV Mat class (n-dimensional dense array class). The other 1/3 is spent on direct access through OpenCV’s Vec class (short numerical vectors). The for-loop is structured divides the picture up based on brush size. Then it finds the colour for each pixel in that section. Finally, it then averages the intensity to produce the final colour of that group of pixels. This is what makes this program ideal for parallelizing, because each iteration of this for-loop is calculating the final colours for each pixel. (SIMD type of process, the single instruction is to find the final colour and the multiple data is the pixels.) | ||
| Line 352: | Line 364: | ||
---- | ---- | ||
| + | |||
| + | <h4>DNA Edit Distance and Sequence Alignment Tool - Analysis by Jonathan D.</h4> | ||
| + | |||
| + | This program is a bioinformatics tool which will calculate the edit distance between two sequences. Furthermore, if the option is selected, then the program will also perform a sequence alignment on the two sequences. The two sequences must be given in FASTA format, but there are already ready to use sequences within the test directory of the project. | ||
| + | |||
| + | Original source code can be be found [https://github.com/kbiscanic/bioinformatics here]. | ||
| + | |||
| + | <h5>Compilation</h5> | ||
| + | To compile the program, navigate to the project’s src directory and run the following command: | ||
| + | |||
| + | $ g++ -std=c++11 -g -pg -O2 BasicEditDistance.cpp main.cpp Parser.cpp Result.cpp Sequence.cpp Solver.cpp SubmatrixCalculator.cpp Writer.cpp | ||
| + | |||
| + | <h5>Running the Program</h5> | ||
| + | There are three options to run the program, as noted in the github readme: | ||
| + | |||
| + | b - basic edit distance (Needleman-Wunsch) | ||
| + | |||
| + | d - edit distance (Masek-Paterson) | ||
| + | |||
| + | a - edit distance and alignment (Masek-Paterson) | ||
| + | |||
| + | Testing of the program was done by using the "a" option which utilizes the Masek-Paterson algorithm to calculate edit distance and perform alignment sequencing on maximum sequence length of 100 nucleotides. (FASTA files were included in the project under the test directory) | ||
| + | |||
| + | <h6>INPUT FILE (test-100.fa):</h6> | ||
| + | |||
| + | >Chromosome_3565586_3565685_0:0:0_0:0:0_0/1 | ||
| + | ACCGGTTGCCCGCTACATGCTCCAACCATCCGGCGATGGTTACCTGCTGCCGGACTGGTATAGCGCAGAGCCGCGTCGACACCGCGTATCCGTGCCCCCC | ||
| + | >Chromosome_3568561_3568661_0:0:1_0:0:1_1/1 | ||
| + | TGGGGATTGCCAGTCCGTCCGGGGAGGTATTCAGAAAGGTACACCGGTCTGTTGATATTCATGTAACAGGTATTAATGATGAAGAAAGGAATGGCAAACA | ||
| + | |||
| + | |||
| + | Run the following command to align the two sequences: | ||
| + | |||
| + | $ ./a.out a ../test/data/test-100.fa test.maf | ||
| + | |||
| + | <h6>SCREEN OUTPUT:</h6> | ||
| + | <pre> | ||
| + | Submatrix dimension: 1 | ||
| + | |||
| + | String A size: 100 | ||
| + | |||
| + | String B size: 100 | ||
| + | |||
| + | Submatrices in edit table: 100x100 | ||
| + | |||
| + | Allocating 230 locations. | ||
| + | |||
| + | Allocation time: 1e-05s | ||
| + | |||
| + | 1 / 5 (submatrices: 45 ) | ||
| + | |||
| + | 5 / 5 (submatrices: 225 ) | ||
| + | |||
| + | Submatrix calculation time: 8.9e-05s | ||
| + | |||
| + | Edit path calculation (Masek-Paterson): 0.000149 | ||
| + | </pre> | ||
| + | |||
| + | <h6>TEXT OUTPUT (test.maf)</h6> | ||
| + | |||
| + | a score=58 | ||
| + | s Chromosome_3565586_3565685_0:0:0_0:0:0_0/1 0 100 + 107 ACCGG-TTGCCCGCTACATGC-TCCAACCATCCGGCGATGGT-TACCTG-CTGCCG-GACTGGTATAGC--GCAGAGCCGCGTCGACACCGCGTATCCGTGCCCCCC | ||
| + | s Chromosome_3568561_3568661_0:0:1_0:0:1_1/1 0 100 + 107 TGGGGATTGCCAG-TCCGTCCGGGGAGGTATTCAG-AAAGGTACACCGGTCTGTTGATATTCATGTAACAGGTATTAATG-ATGAAGAAAG-GAAT--G-GCAAACA | ||
| + | |||
| + | As you can see, the two sequences are now aligned which will show conserved regions of nucleotides between the two. | ||
| + | |||
| + | <h5>Analysis</h5> | ||
| + | |||
| + | In order to perform gprof analysis, sequence length of 10000 was used: | ||
| + | |||
| + | $ ./a.out a ../test/data/test-10000.fa test.maf | ||
| + | $ gprof -p -b > FULL_O2.analysis.10000.flt | ||
| + | |||
| + | Flat profile: | ||
| + | Each sample counts as 0.01 seconds. | ||
| + | % cumulative self self total | ||
| + | time seconds seconds calls s/call s/call name | ||
| + | 36.59 0.45 0.45 302086997 0.00 0.00 std::vector<int, std::allocator<int> >::operator[](unsigned long) | ||
| + | 22.77 0.73 0.28 1 0.28 1.05 Solver::fill_edit_matrix() | ||
| + | 13.82 0.90 0.17 201301730 0.00 0.00 std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > >::operator[](unsigned long) | ||
| + | 12.20 1.05 0.15 25000000 0.00 0.00 SubmatrixCalculator::sumSteps(int) | ||
| + | 7.32 1.14 0.09 727566 0.00 0.00 std::vector<int, std::allocator<int> >::size() const | ||
| + | 3.25 1.18 0.04 5 0.01 0.01 std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > >::vector() | ||
| + | 0.81 1.19 0.01 284812 0.00 0.00 std::_Vector_base<int, std::allocator<int> >::_M_deallocate(int*, unsigned long) | ||
| + | |||
| + | Even with a low sequence length of 10000, we can already begin to see that a lot of time is used in the function call Solver::fill_edit_matrix(). | ||
| + | |||
| + | If we run the program for a larger sequence of 50000, the following flat profile is generated. | ||
| + | |||
| + | Flat profile: | ||
| + | Each sample counts as 0.01 seconds. | ||
| + | % cumulative self self total | ||
| + | time seconds seconds calls s/call s/call name | ||
| + | 85.67 5.61 5.61 1 5.61 5.61 Solver::fill_edit_matrix() | ||
| + | 8.09 6.14 0.53 5267025 0.00 0.00 SubmatrixCalculator::calculateFinalSteps(std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >) | ||
| + | 3.05 6.34 0.20 63261681 0.00 0.00 std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > >::_M_fill_insert(__gnu_cxx::__normal_iterator<std::vector<int, std::allocator<int> >*, std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > > >, unsigned long, std::vector<int, std::allocator<int> > const&) | ||
| + | 3.05 6.54 0.20 1 0.20 0.93 SubmatrixCalculator::calculate() | ||
| + | |||
| + | At 50000 nucleotides, it is even more clear that the Solver::fill_edit_matrix() function is the dominating term of the execution since it's taking up 85% of the elapsed time. | ||
| + | |||
| + | <h6>Code Snippet</h6> | ||
| + | |||
| + | <pre> | ||
| + | |||
| + | /* | ||
| + | Backtracks through the submatrices until it reaches the | ||
| + | starting cell. Edit operations in the vector will be ordered backwards | ||
| + | and the integers represent: | ||
| + | 1 - moving down in the submatrix | ||
| + | 2 - moving right in the submatrix | ||
| + | 3 - moving diagonally in the submatrix | ||
| + | */ | ||
| + | |||
| + | void Solver::fill_edit_matrix() { | ||
| + | all_columns.resize(row_num + 1, vector<int>(column_num + 1, 0)); | ||
| + | all_rows.resize(row_num + 1, vector<int>(column_num + 1, 0)); | ||
| + | top_left_costs.resize(row_num + 1, vector<int>(column_num + 1, 0)); | ||
| + | |||
| + | int initialVector = 0; | ||
| + | for (int i = 0; i < submatrix_dim; i++){ | ||
| + | initialVector = initialVector * 10 + 2; // 222 == "222" == (1, 1, 1) | ||
| + | } | ||
| + | |||
| + | // padding string b step vectors | ||
| + | for (int submatrix_j = 1; submatrix_j <= column_num; submatrix_j++) { | ||
| + | if ((submatrix_j * submatrix_dim - 1) >= string_b_real_size) { | ||
| + | vector<int> temp_vec(submatrix_dim, 0); | ||
| + | for (int i = 0; | ||
| + | i < (string_b_real_size - ((submatrix_j - 1) * submatrix_dim)); | ||
| + | i++) | ||
| + | temp_vec[i] = 1; | ||
| + | all_rows[0][submatrix_j] = SubmatrixCalculator::stepsToInt(temp_vec); | ||
| + | } else { | ||
| + | all_rows[0][submatrix_j] = initialVector; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | // padding string a step vectors | ||
| + | for (int submatrix_i = 1; submatrix_i <= row_num; submatrix_i++) { | ||
| + | if ((submatrix_i * submatrix_dim - 1) >= string_a_real_size) { | ||
| + | vector<int> temp_vec(submatrix_dim, 0); | ||
| + | for (int i = 0; | ||
| + | i < (string_a_real_size - ((submatrix_i - 1) * submatrix_dim)); | ||
| + | i++) | ||
| + | temp_vec[i] = 1; | ||
| + | all_columns[submatrix_i][0] = | ||
| + | SubmatrixCalculator::stepsToInt(temp_vec); | ||
| + | } else { | ||
| + | all_columns[submatrix_i][0] = initialVector; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | for (int submatrix_i = 1; submatrix_i <= row_num; submatrix_i++) { | ||
| + | if ((submatrix_i - 1) * submatrix_dim > string_a_real_size) | ||
| + | top_left_costs[submatrix_i][1] = string_a_real_size; | ||
| + | else | ||
| + | top_left_costs[submatrix_i][1] = (submatrix_i - 1) * submatrix_dim; | ||
| + | |||
| + | for (int submatrix_j = 1; submatrix_j <= column_num; submatrix_j++) { | ||
| + | |||
| + | pair<int, int> final_steps = subm_calc->resultIndex[ | ||
| + | // offset calculation | ||
| + | str_a_offsets[submatrix_i] + // left string | ||
| + | str_b_offsets[submatrix_j] + // top string | ||
| + | // left steps | ||
| + | subm_calc->stepOffsets[0][all_columns[submatrix_i][submatrix_j - 1]] + | ||
| + | // top steps | ||
| + | subm_calc->stepOffsets[1][all_rows[submatrix_i - 1][submatrix_j]] | ||
| + | ]; | ||
| + | |||
| + | all_columns[submatrix_i][submatrix_j] = final_steps.first; | ||
| + | all_rows[submatrix_i][submatrix_j] = final_steps.second; | ||
| + | |||
| + | if (submatrix_j != 1) { | ||
| + | top_left_costs[submatrix_i][submatrix_j] = | ||
| + | top_left_costs[submatrix_i][submatrix_j - 1]; | ||
| + | top_left_costs[submatrix_i][submatrix_j] += subm_calc->sumSteps( | ||
| + | all_rows[submatrix_i - 1][submatrix_j - 1]); | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | Within the function itself there are many nested loops with calls to the sumSteps function in the SubmatrixCalculator.cpp file. Since the purpose of this function is to retrieve the path through the matrix while calculating the number of steps, this simple operation could benefit from the usage of a GPU rather than a CPU. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | <h4>Recommendation</h4> | ||
| + | |||
| + | Based on the above profiling, our group has decided to go with Sudoku solver as our program to parallelize. | ||
| + | |||
| + | In order to determine the potential speed up for the Sudoku solver, we utilized Amdahl's Law: | ||
| + | |||
| + | S480 = 1 / ( 1 - 0.47 + 0.47 / 480 ) = 1.88 | ||
| + | |||
| + | At best, we expect the process time to drop from 17.13 secs to about 17.13 / 1.88 = 9.11 secs | ||
| + | |||
| + | CheckSquare() | ||
| + | O(n) = 8n^3 + 3n^2 + 10n | ||
| + | |||
| + | CheckColumn() | ||
| + | O(n) = 6n^2 + 4n | ||
| + | |||
| + | CheckRow() | ||
| + | O(n) = 6n^2 + 4n | ||
| + | |||
| + | Print() | ||
| + | O(n) = 4n^3 + 3n^2 + 3n | ||
| + | |||
| + | StorePositions() | ||
| + | O(n) = 4n^3 + 3n^2 + 3n | ||
| + | |||
| + | GoBack() | ||
| + | O(n) = 11n | ||
| + | |||
| + | PlaceNum() | ||
| + | O(n) = 7n^2 + 5n | ||
| + | |||
| + | SolveSudoku() | ||
| + | O(n) = 8n^3 + 3n^2 + 2n | ||
| + | |||
| + | Main() | ||
| + | O(n) = 15n^3 + 7n^2 + 15n | ||
| + | |||
| + | Entire program | ||
| + | O(n) = 42n^3 + 38n^2 + 59n | ||
| + | |||
| + | The Big-O of the Sudoku program is O(n^3), or cubic. This program would be idea for parallalizing. | ||
| + | |||
| + | The reason we chose the Sudoku solver was because we felt that the Sequence Alignment was too complex of an algorithm to work with, and the image processor seemed a bit too simple to parallelize. The sudoku solver seemed to be the right balance in terms of complexity as well as potential speed up. We are nearly cutting the time in half for a standard 9x9 puzzle, but as the puzzles get harder, through either an increase in the matrix size, or through an increase in blank numbers to be solved, we would expect even better performance through parallel programming than standard serial programming. | ||
=== Assignment 2 === | === Assignment 2 === | ||
| + | <h4>Parallelized Oil Painting Program</h4> | ||
| + | Despite our initial decision of choosing to parallelize the Sudoku solver in assignment 1, we came to the conclusion that parallelizing the oil painting program would be better suited for us. We were able to grasp the logic behind the oil painting program whereas we had a lot of trouble working out the logic behind solving Sudoku puzzles. | ||
| + | |||
| + | <h4>The Code</h4> | ||
| + | [[Media: A2-Blastoise.zip]] This is a zip file that contains our full code and an executable version. To create your own project in visual studio with this code, you will need to download OpenCV. You will need the following property settings in your project: add the include path of OpenCV to VC++ Directories -> Include Directories and also add opencv_world320d.lib to Linker -> Input and add a post build command to copy OpenCV dll files into your project directory. (The last step can also be done by copying OpenCV bin files into debug directory of your project.) | ||
| + | |||
| + | In the serial program, the oil painting worked by going one pixel at a time in a double for loop. | ||
| + | |||
| + | <pre> | ||
| + | for (int i=0; i < height; i++) | ||
| + | { | ||
| + | for (int j=0; j < width; j++) | ||
| + | { | ||
| + | //pixel processing | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | We were able to remove the need for this loop through the utilization of a kernel. In the main function, we created the following block and grid and called our oil painting kernel. The ceil function was used to calculate the grid dimensions, so that based on the block size the entire image will be included in the block. | ||
| + | |||
| + | <pre> | ||
| + | const dim3 block(ntpb, ntpb); // ntpb was calculated based on device property (maxThreadsDim). | ||
| + | const dim3 grid(ceil((float)width / block.x), ceil((float)height / block.y), 1); | ||
| + | oilPaint << <grid, block >> >(gpu_src, gpu_dst, width, height); | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | In our kernel, through the use of the primitive types, we are able to determine the exact position of the pixel in the 2D array. Now instead of iterating through every pixel in the image, each thread in the block will adjust its own pixel intensity. The overall logic of the code stayed the same. We moved the double for loop into kernel and used i and j to locate the pixel. | ||
| + | |||
| + | <pre> | ||
| + | //2D Index of current thread | ||
| + | const int i = blockIdx.x * blockDim.x + threadIdx.x; | ||
| + | const int j = blockIdx.y * blockDim.y + threadIdx.y; | ||
| + | </pre> | ||
| + | |||
| + | <h4>The Results</h4> | ||
| + | The first graph is a comparison of the original execution time and the parallelize version. There was a considerable speed up. The original seems to have a growth rate that is exponential while the parallelized version is practically logarithmic. The second graph shows the time spent in the kernel in milliseconds, while also showing the percentage of time spend on the device verses host. The time spent in the kernel increases with the problem size, as expected. You can see that the time spent on the device also has a growth rate that is logarithmic, this means that for extremely small problem sizes the parallel version might not have a great speed up. (This is caused by the amount of CUDA API calls.) | ||
| + | |||
| + | [[File:A2-Result.PNG]] | ||
| + | |||
=== Assignment 3 === | === Assignment 3 === | ||
| + | |||
| + | [[Media: OilPaint.zip]] This is our complete optimized solution, including the executable, code, and an image. The original parallelized solution took 3 arguments, the brush size, intensity level, the file name. Our default brush size was 5, and intensity level was 20, these values would be good for testing. Our optimized solution took only 1 argument, the file name. To run the parallel and optimized examples, the OpenCV files, specifically opencv_world320d.dll needs to be in the same directory as the executable. We tried multiple optimization method, some of them worked while others made our times worse. Below we have all our attempts at optimizations, what worked and what made it worse. | ||
| + | |||
| + | <h4>Shared memory</h4> | ||
| + | |||
| + | We attempted two potential applications of shared memory. Our first try was to store the entire image in shared memory. We realized that there was no need because this source image was only accessed once in the algorithm. Therefore transferring the image from global to shared, and then back to global, would be very inefficient. To make shared memory efficient, we would need to access it multiple times for any actual speedup. | ||
| + | |||
| + | |||
| + | Our second attempt was to store the array needed to calculate intensity. We realize that this would not be possible as each thread would require their own colour value arrays for calculations done at different times | ||
| + | |||
| + | |||
| + | <h4>Constant memory</h4> | ||
| + | We declared our image as constant memory. Constant memory was implemented and that increased our speed. However because the size of the image is dynamic, this can't be considered real constant memory. | ||
| + | |||
| + | |||
| + | <h4>Instruction mixing</h4> | ||
| + | For instruction mixing, we unrolled the inner loop with a constant brush size. We removed the option for the user to change the brush size and unrolled the loop with a constant brush size. We have so many checks and other functions inside of our inner loop that unrolling was causing it to slow down slightly. The result was a slight speed down and therefore we did not use instruction mixing. However we kept some functions that slightly sped the processing up. | ||
| + | |||
| + | |||
| + | <h4>Reduce CUDA API calls</h4> | ||
| + | We attempted to use only 1 array to hold both the source and destination. Because the destination image calculations depend on data from the source, we cannot change the source as we go. | ||
| + | |||
| + | |||
| + | <h4>Coalesced Access</h4> | ||
| + | Our last attempt at optimization was to try using coalesced access. We switched the ''x'' and ''y'' grid values so that all the values are adjacent to each other in memory. This resulted in a 1.3x speedup when combined with constant memory and occupancy optimization. | ||
| + | |||
| + | |||
| + | |||
| + | <h4>Optimization results</h4> | ||
| + | |||
| + | [[File:Capture.PNG]] | ||
| + | |||
| + | These are our results after different optimization steps. The combination of our optimizations resulted in a 1.3x speedup compared to our un-optimized version. | ||
Latest revision as of 09:32, 13 April 2017
GPU610/DPS915 | Student List | Group and Project Index | Student Resources | Glossary
Contents
Project Name Goes here
Team Members
- Matt Babol, Sudoku
- Jonathan Desmond, DNA Edit Distance and Alignment Tool
- Sallie Jiang, Oil Painting
_
_,..-"""--' `,.-".
,' __.. --', |
_/ _.-"' | .' | | ____
,.-""' `-"+.._| `.' | `-..,',--.`.
| ,. ' j 7 l \__
|.-' /| | j|| .
`. | / L`.`""','|\ \
`.,----..._ ,'`"'-. ,' \ `""' | | l
Y `-----' v' ,'`,.__..' | .
`. / / / `.| |
`. / l j ,^. |L
`._ L +. |._ .' \| | \
.`--...__,..-'""'-._ l L """ | | \
.' ,`-......L_ \ \ \ _.' ,'. l
,-"`. / ,-.---.' `. \ L..--"' _.-^.| l
.-"".'"`. Y `._' ' `. | | _,.--'" | |
`._' | |,-'| l `. | |".. | l
,'. | |`._' | `. | |_,...---"""""` L
/ | j _|-' `. L | j ,| | |
`-,"._,-+' /`---^..../._____,.L',' `. |\ |
|,' L | `-. | \j
. \ `, | |
\ __`.Y._ -. j |
\ _.,' `._ \ | j
,-"`-----""""' |`. \ 7 |
/ `. ' | \ \ / |
| ` / | \ Y |
| \ . ,' | L_.-')
L `. | / ] _.-^._
\ ,' `-7 ,-' / | ,' `-._
_,`._ `. _,-' ,',^.- `.
,-' v.... _.`"', _:'--....._______,.-'
._______./ /',,-'"'`'--. ,-' `.
"""""`.,' _\`----...'
--------""'
Progress
Assignment 1
Sudoku Solver by Matt B.
Sudoku solver is a program that solves a sudoku puzzle. The user can input an existing file and have the program solve this, or can manually enter values to be solved. The sudoku puzzle is 9x9 in size. The data needs to be in a specific format for the program to work. There are 9 rows of values, with each cell/element in the row needing to be separated by a space. A value of 0 tells the program to solve this value.
Original source code can be found here.
Easy puzzle
Running program
To compile the program, open the terminal and go to the projects directory
$ g++ -std=c++0x -pg solver.cpp checks.cpp checksolution.cpp -o Sudoku
This will create an executable file called Sudoku. -pg is used for creating a gmon.out file, which will allow us to profile the program with arguments.
This is the easy sudoku puzzle that will be running through the program first. The file is saved as 'puzzle' in the same directory.
0 6 0 0 0 0 9 7 2 0 5 0 0 0 2 0 0 3 0 7 0 3 9 0 5 0 0 2 0 0 0 0 5 4 0 8 0 0 0 0 0 0 0 0 0 3 0 1 8 0 0 0 0 6 0 0 4 0 2 3 0 8 0 7 0 0 9 0 0 0 2 0 9 2 5 0 0 0 0 4 0
Run the code with
$ ./Sudoku puzzle
After the program is done running, the result is
1 6 3 4 5 8 9 7 2 4 5 9 7 1 2 8 6 3 8 7 2 3 9 6 5 1 4 2 9 7 1 6 5 4 3 8 5 8 6 2 3 4 1 9 7 3 4 1 8 7 9 2 5 6 6 1 4 5 2 3 7 8 9 7 3 8 9 4 1 6 2 5 9 2 5 6 8 7 3 4 1
Test Case
To profile the program, run this command.
$ gprof -p -b ./Sudoku gmon.out > Sudoku.flt
The profiling result
Flat profile: Each sample counts as 0.01 seconds. no time accumulated % cumulative self self total time seconds seconds calls Ts/call Ts/call name 0.00 0.00 0.00 4539 0.00 0.00 checkRow(int, int) 0.00 0.00 0.00 1620 0.00 0.00 checkColumn(int, int) 0.00 0.00 0.00 1120 0.00 0.00 placeNum(int, int) 0.00 0.00 0.00 698 0.00 0.00 checkSquare(int, int, int) 0.00 0.00 0.00 476 0.00 0.00 goBack(int&, int&) 0.00 0.00 0.00 2 0.00 0.00 print(int (*) [9]) 0.00 0.00 0.00 1 0.00 0.00 _GLOBAL__sub_I_sudoku 0.00 0.00 0.00 1 0.00 0.00 _GLOBAL__sub_I_temp 0.00 0.00 0.00 1 0.00 0.00 solveSudoku() 0.00 0.00 0.00 1 0.00 0.00 storePositions() 0.00 0.00 0.00 1 0.00 0.00 __static_initialization_and_destruction_0(int, int) 0.00 0.00 0.00 1 0.00 0.00 __static_initialization_and_destruction_0(int, int)
Call graph
granularity: each sample hit covers 2 byte(s) no time propagated
index % time self children called name
0.00 0.00 4539/4539 placeNum(int, int) [10]
[8] 0.0 0.00 0.00 4539 checkRow(int, int) [8]
-----------------------------------------------
0.00 0.00 1620/1620 placeNum(int, int) [10]
[9] 0.0 0.00 0.00 1620 checkColumn(int, int) [9]
-----------------------------------------------
0.00 0.00 1120/1120 solveSudoku() [16]
[10] 0.0 0.00 0.00 1120 placeNum(int, int) [10]
0.00 0.00 4539/4539 checkRow(int, int) [8]
0.00 0.00 1620/1620 checkColumn(int, int) [9]
0.00 0.00 698/698 checkSquare(int, int, int) [11]
-----------------------------------------------
0.00 0.00 698/698 placeNum(int, int) [10]
[11] 0.0 0.00 0.00 698 checkSquare(int, int, int) [11]
-----------------------------------------------
0.00 0.00 476/476 solveSudoku() [16]
[12] 0.0 0.00 0.00 476 goBack(int&, int&) [12]
-----------------------------------------------
0.00 0.00 2/2 main [6]
[13] 0.0 0.00 0.00 2 print(int (*) [9]) [13]
-----------------------------------------------
0.00 0.00 1/1 __libc_csu_init [30]
[14] 0.0 0.00 0.00 1 _GLOBAL__sub_I_sudoku [14]
0.00 0.00 1/1 __static_initialization_and_destruction_0(int, int) [18]
-----------------------------------------------
0.00 0.00 1/1 __libc_csu_init [30]
[15] 0.0 0.00 0.00 1 _GLOBAL__sub_I_temp [15]
0.00 0.00 1/1 __static_initialization_and_destruction_0(int, int) [19]
-----------------------------------------------
0.00 0.00 1/1 main [6]
[16] 0.0 0.00 0.00 1 solveSudoku() [16]
0.00 0.00 1120/1120 placeNum(int, int) [10]
0.00 0.00 476/476 goBack(int&, int&) [12]
-----------------------------------------------
0.00 0.00 1/1 main [6]
[17] 0.0 0.00 0.00 1 storePositions() [17]
-----------------------------------------------
0.00 0.00 1/1 _GLOBAL__sub_I_sudoku [14]
[18] 0.0 0.00 0.00 1 __static_initialization_and_destruction_0(int, int) [18]
-----------------------------------------------
0.00 0.00 1/1 _GLOBAL__sub_I_temp [15]
[19] 0.0 0.00 0.00 1 __static_initialization_and_destruction_0(int, int) [19]
-----------------------------------------------
Index by function name
[14] _GLOBAL__sub_I_sudoku [16] solveSudoku() [13] print(int (*) [9])
[15] _GLOBAL__sub_I_temp [17] storePositions() [12] goBack(int&, int&)
[9] checkColumn(int, int) [18] __static_initialization_and_destruction_0(int, int) [8] checkRow(int, int)
[11] checkSquare(int, int, int) [19] __static_initialization_and_destruction_0(int, int) [10] placeNum(int, int)
Analysis
The analysis reveals that the program took no time in solving this puzzle. However, the function checkRow and checkColumn were called the most. These two functions are used for checking whether the row and columns are correct. For a deeper analysis, a harder puzzle must be used.
Hard puzzle
Running program
For the hard puzzle, below is the input file as well as the result
0 0 0 0 0 0 0 0 0 0 0 0 0 0 3 0 8 5 0 0 1 0 2 0 0 0 0 0 0 0 5 0 7 0 0 0 0 0 4 0 0 0 1 0 0 0 9 0 0 0 0 0 0 0 5 0 0 0 0 0 0 7 3 0 0 2 0 1 0 0 0 0 0 0 0 0 4 0 0 0 9 9 8 7 6 5 4 3 2 1 2 4 6 1 7 3 9 8 5 3 5 1 9 2 8 7 4 6 1 2 8 5 3 7 6 9 4 6 3 4 8 9 2 1 5 7 7 9 5 4 6 1 8 3 2 5 1 9 2 8 6 4 7 3 4 7 2 3 1 9 5 6 8 8 6 3 7 4 5 2 1 9
Test Cases
The profiling results are
Flat profile: Each sample counts as 0.01 seconds. % cumulative self self total time seconds seconds calls s/call s/call name 45.34 7.76 7.76 622577597 0.00 0.00 checkRow(int, int) 18.63 10.94 3.19 223365661 0.00 0.00 checkColumn(int, int) 14.38 13.40 2.46 157353814 0.00 0.00 placeNum(int, int) 13.24 15.67 2.27 100608583 0.00 0.00 checkSquare(int, int, int) 4.80 16.49 0.82 69175252 0.00 0.00 goBack(int&, int&) 3.46 17.08 0.59 1 0.59 17.08 solveSudoku() 0.29 17.13 0.05 1 0.05 0.05 _GLOBAL__sub_I_sudoku 0.00 17.13 0.00 2 0.00 0.00 print(int (*) [9]) 0.00 17.13 0.00 1 0.00 0.00 _GLOBAL__sub_I_temp 0.00 17.13 0.00 1 0.00 0.00 storePositions() 0.00 17.13 0.00 1 0.00 0.00 __static_initialization_and_destruction_0(int, int) 0.00 17.13 0.00 1 0.00 0.00 __static_initialization_and_destruction_0(int, int)
Call graph
granularity: each sample hit covers 2 byte(s) for 0.06% of 17.13 seconds
index % time self children called name
<spontaneous>
[1] 99.7 0.00 17.08 main [1]
0.59 16.49 1/1 solveSudoku() [2]
0.00 0.00 2/2 print(int (*) [9]) [16]
0.00 0.00 1/1 storePositions() [18]
-----------------------------------------------
0.59 16.49 1/1 main [1]
[2] 99.7 0.59 16.49 1 solveSudoku() [2]
2.46 13.21 157353814/157353814 placeNum(int, int) [3]
0.82 0.00 69175252/69175252 goBack(int&, int&) [7]
-----------------------------------------------
2.46 13.21 157353814/157353814 solveSudoku() [2]
[3] 91.5 2.46 13.21 157353814 placeNum(int, int) [3]
7.76 0.00 622577597/622577597 checkRow(int, int) [4]
3.19 0.00 223365661/223365661 checkColumn(int, int) [5]
2.27 0.00 100608583/100608583 checkSquare(int, int, int) [6]
-----------------------------------------------
7.76 0.00 622577597/622577597 placeNum(int, int) [3]
[4] 45.3 7.76 0.00 622577597 checkRow(int, int) [4]
-----------------------------------------------
3.19 0.00 223365661/223365661 placeNum(int, int) [3]
[5] 18.6 3.19 0.00 223365661 checkColumn(int, int) [5]
-----------------------------------------------
2.27 0.00 100608583/100608583 placeNum(int, int) [3]
[6] 13.2 2.27 0.00 100608583 checkSquare(int, int, int) [6]
-----------------------------------------------
0.82 0.00 69175252/69175252 solveSudoku() [2]
[7] 4.8 0.82 0.00 69175252 goBack(int&, int&) [7]
-----------------------------------------------
0.05 0.00 1/1 __libc_csu_init [9]
[8] 0.3 0.05 0.00 1 _GLOBAL__sub_I_sudoku [8]
0.00 0.00 1/1 __static_initialization_and_destruction_0(int, int) [19]
-----------------------------------------------
<spontaneous>
[9] 0.3 0.00 0.05 __libc_csu_init [9]
0.05 0.00 1/1 _GLOBAL__sub_I_sudoku [8]
0.00 0.00 1/1 _GLOBAL__sub_I_temp [17]
-----------------------------------------------
0.00 0.00 2/2 main [1]
[16] 0.0 0.00 0.00 2 print(int (*) [9]) [16]
-----------------------------------------------
0.00 0.00 1/1 __libc_csu_init [9]
[17] 0.0 0.00 0.00 1 _GLOBAL__sub_I_temp [17]
0.00 0.00 1/1 __static_initialization_and_destruction_0(int, int) [20]
-----------------------------------------------
0.00 0.00 1/1 main [1]
[18] 0.0 0.00 0.00 1 storePositions() [18]
-----------------------------------------------
0.00 0.00 1/1 _GLOBAL__sub_I_sudoku [8]
[19] 0.0 0.00 0.00 1 __static_initialization_and_destruction_0(int, int) [19]
-----------------------------------------------
0.00 0.00 1/1 _GLOBAL__sub_I_temp [17]
[20] 0.0 0.00 0.00 1 __static_initialization_and_destruction_0(int, int) [20]
-----------------------------------------------
Index by function name
[8] _GLOBAL__sub_I_sudoku [2] solveSudoku() [16] print(int (*) [9])
[17] _GLOBAL__sub_I_temp [18] storePositions() [7] goBack(int&, int&)
[5] checkColumn(int, int) [19] __static_initialization_and_destruction_0(int, int) [4] checkRow(int, int)
[6] checkSquare(int, int, int) [20] __static_initialization_and_destruction_0(int, int) [3] placeNum(int, int)
Analysis
With a harder puzzle, the time for the program to solve the puzzle increased significantly. The total time to complete this puzzle was 17.13 seconds. The program spends almost half of its running time checking if the row is correct, and another 18% of its time checking whether the column is correct. This program contains thousands of calls to check the row and column values, this is why the program would be excellent project for parallelizing.
Oil Painting By Sallie J.
This program converts a regular image into a stylized oil painting, it uses OpenCV. The painting algorithm depends on the brush size and colour intensity. The program takes three command line arguments: int for brush size, int for intensity and file name of an image. Upon finishing, the program produces the original image along with the oil paint version and the total time required in seconds.
The original source code can be found here.
However there have been some changes to make testing and profiling slightly easier. (Mainly changes are putting the for-loop logic into a function outside of the main and modifying for command line arguments instead of hard coding values.)
Running the program
To compile the program on Linux you must download the OpenCV library and then create a makefile that will create the executable OilPaint.exe. To compile the program in visual studio you will need to set project properties for OpenCV through setting the C/C++ Additional Include Directories, Linker Additional Library Directories and Input Additional Dependencies (opencv_world320d.lib).
Media: OilPaint.zip This zip file contains a make file and the cpp file. To compile on Linux alter the makefile. Run the executable with the arguments 5 (brush size), 20 (colour intensity), filename.format (including file format)
$ cmake -DCMAKE_CXX_FLAGS=-pg =DCMAKE_EXE_LINKER_FLAGS=-pg -DCMAKE_SHARED_LINKER_FLAGS=-pg -pg . $ make $ ./OilPaint 5 20 filename.fileformat
Output:
gprof Output:
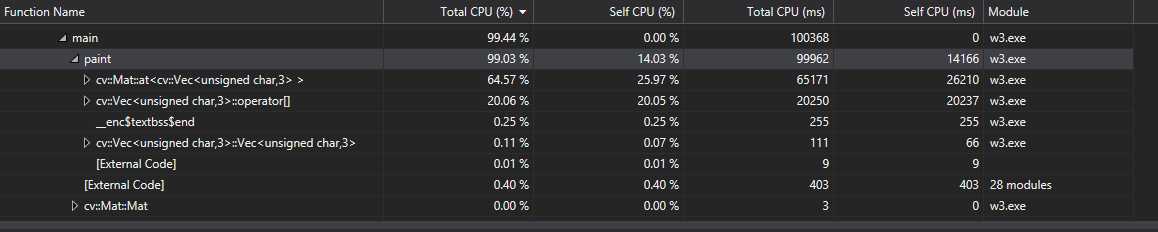
Flat profile: Each sample counts as 0.01 seconds. % cumulative self self total time seconds seconds calls s/call s/call name 79.79 20.17 20.17 1 20.17 24.77 paint(cv::Mat&, cv::Mat&, int, int, int, int) 12.53 23.34 3.17 1742140400 0.00 0.00 cv::Vec<unsigned char, 3>& cv::Mat::at<cv::Vec<unsigned char, 3> >(int, int) 5.62 24.76 1.42 1737354300 0.00 0.00 cv::Vec<unsigned char, 3>::operator[](int) 1.94 25.25 0.49 1 0.49 0.49 cv::Size_<int>::Size_(int, int) ... (there are a lot of other calls to the library that did not significantly affect the profiling)
Visual Studio Performance profiler Output:
The time required for the program depends largely on the file size being converted. Around 5 seconds for a 50KB image and 100 seconds for a 1MB image. It depends on the brush size and intensity levels as well.
Analysis
The profiling revealed that 80-99% of the processing time is spent in the paint function where the for-loop logic is located. Within that 99% the program spends roughly 2/3 of its time reading accessing data through the "at" function of the OpenCV Mat class (n-dimensional dense array class). The other 1/3 is spent on direct access through OpenCV’s Vec class (short numerical vectors). The for-loop is structured divides the picture up based on brush size. Then it finds the colour for each pixel in that section. Finally, it then averages the intensity to produce the final colour of that group of pixels. This is what makes this program ideal for parallelizing, because each iteration of this for-loop is calculating the final colours for each pixel. (SIMD type of process, the single instruction is to find the final colour and the multiple data is the pixels.)
//Simplified for-loop structure
for (int y = BrushSize; y < (height - BrushSize); y++) //for each row based on brush size
{
for (int x = BrushSize; x < (width - BrushSize); x++) //for each column in brush size
{
for (int j = -BrushSize; j <= BrushSize; j++) //for each pixel row in one brush size grouping
{
for (int i = -BrushSize; i <= BrushSize; i++)//for each pixel column in one brush size grouping
{ //algorithm and logic for colour calculations }
}
}
}
DNA Edit Distance and Sequence Alignment Tool - Analysis by Jonathan D.
This program is a bioinformatics tool which will calculate the edit distance between two sequences. Furthermore, if the option is selected, then the program will also perform a sequence alignment on the two sequences. The two sequences must be given in FASTA format, but there are already ready to use sequences within the test directory of the project.
Original source code can be be found here.
Compilation
To compile the program, navigate to the project’s src directory and run the following command:
$ g++ -std=c++11 -g -pg -O2 BasicEditDistance.cpp main.cpp Parser.cpp Result.cpp Sequence.cpp Solver.cpp SubmatrixCalculator.cpp Writer.cpp
Running the Program
There are three options to run the program, as noted in the github readme:
b - basic edit distance (Needleman-Wunsch)
d - edit distance (Masek-Paterson)
a - edit distance and alignment (Masek-Paterson)
Testing of the program was done by using the "a" option which utilizes the Masek-Paterson algorithm to calculate edit distance and perform alignment sequencing on maximum sequence length of 100 nucleotides. (FASTA files were included in the project under the test directory)
INPUT FILE (test-100.fa):
>Chromosome_3565586_3565685_0:0:0_0:0:0_0/1 ACCGGTTGCCCGCTACATGCTCCAACCATCCGGCGATGGTTACCTGCTGCCGGACTGGTATAGCGCAGAGCCGCGTCGACACCGCGTATCCGTGCCCCCC >Chromosome_3568561_3568661_0:0:1_0:0:1_1/1 TGGGGATTGCCAGTCCGTCCGGGGAGGTATTCAGAAAGGTACACCGGTCTGTTGATATTCATGTAACAGGTATTAATGATGAAGAAAGGAATGGCAAACA
Run the following command to align the two sequences:
$ ./a.out a ../test/data/test-100.fa test.maf
SCREEN OUTPUT:
Submatrix dimension: 1 String A size: 100 String B size: 100 Submatrices in edit table: 100x100 Allocating 230 locations. Allocation time: 1e-05s 1 / 5 (submatrices: 45 ) 5 / 5 (submatrices: 225 ) Submatrix calculation time: 8.9e-05s Edit path calculation (Masek-Paterson): 0.000149
TEXT OUTPUT (test.maf)
a score=58 s Chromosome_3565586_3565685_0:0:0_0:0:0_0/1 0 100 + 107 ACCGG-TTGCCCGCTACATGC-TCCAACCATCCGGCGATGGT-TACCTG-CTGCCG-GACTGGTATAGC--GCAGAGCCGCGTCGACACCGCGTATCCGTGCCCCCC s Chromosome_3568561_3568661_0:0:1_0:0:1_1/1 0 100 + 107 TGGGGATTGCCAG-TCCGTCCGGGGAGGTATTCAG-AAAGGTACACCGGTCTGTTGATATTCATGTAACAGGTATTAATG-ATGAAGAAAG-GAAT--G-GCAAACA
As you can see, the two sequences are now aligned which will show conserved regions of nucleotides between the two.
Analysis
In order to perform gprof analysis, sequence length of 10000 was used:
$ ./a.out a ../test/data/test-10000.fa test.maf $ gprof -p -b > FULL_O2.analysis.10000.flt
Flat profile: Each sample counts as 0.01 seconds. % cumulative self self total time seconds seconds calls s/call s/call name 36.59 0.45 0.45 302086997 0.00 0.00 std::vector<int, std::allocator<int> >::operator[](unsigned long) 22.77 0.73 0.28 1 0.28 1.05 Solver::fill_edit_matrix() 13.82 0.90 0.17 201301730 0.00 0.00 std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > >::operator[](unsigned long) 12.20 1.05 0.15 25000000 0.00 0.00 SubmatrixCalculator::sumSteps(int) 7.32 1.14 0.09 727566 0.00 0.00 std::vector<int, std::allocator<int> >::size() const 3.25 1.18 0.04 5 0.01 0.01 std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > >::vector() 0.81 1.19 0.01 284812 0.00 0.00 std::_Vector_base<int, std::allocator<int> >::_M_deallocate(int*, unsigned long)
Even with a low sequence length of 10000, we can already begin to see that a lot of time is used in the function call Solver::fill_edit_matrix().
If we run the program for a larger sequence of 50000, the following flat profile is generated.
Flat profile: Each sample counts as 0.01 seconds. % cumulative self self total time seconds seconds calls s/call s/call name 85.67 5.61 5.61 1 5.61 5.61 Solver::fill_edit_matrix() 8.09 6.14 0.53 5267025 0.00 0.00 SubmatrixCalculator::calculateFinalSteps(std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >) 3.05 6.34 0.20 63261681 0.00 0.00 std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > >::_M_fill_insert(__gnu_cxx::__normal_iterator<std::vector<int, std::allocator<int> >*, std::vector<std::vector<int, std::allocator<int> >, std::allocator<std::vector<int, std::allocator<int> > > > >, unsigned long, std::vector<int, std::allocator<int> > const&) 3.05 6.54 0.20 1 0.20 0.93 SubmatrixCalculator::calculate()
At 50000 nucleotides, it is even more clear that the Solver::fill_edit_matrix() function is the dominating term of the execution since it's taking up 85% of the elapsed time.
Code Snippet
/*
Backtracks through the submatrices until it reaches the
starting cell. Edit operations in the vector will be ordered backwards
and the integers represent:
1 - moving down in the submatrix
2 - moving right in the submatrix
3 - moving diagonally in the submatrix
*/
void Solver::fill_edit_matrix() {
all_columns.resize(row_num + 1, vector<int>(column_num + 1, 0));
all_rows.resize(row_num + 1, vector<int>(column_num + 1, 0));
top_left_costs.resize(row_num + 1, vector<int>(column_num + 1, 0));
int initialVector = 0;
for (int i = 0; i < submatrix_dim; i++){
initialVector = initialVector * 10 + 2; // 222 == "222" == (1, 1, 1)
}
// padding string b step vectors
for (int submatrix_j = 1; submatrix_j <= column_num; submatrix_j++) {
if ((submatrix_j * submatrix_dim - 1) >= string_b_real_size) {
vector<int> temp_vec(submatrix_dim, 0);
for (int i = 0;
i < (string_b_real_size - ((submatrix_j - 1) * submatrix_dim));
i++)
temp_vec[i] = 1;
all_rows[0][submatrix_j] = SubmatrixCalculator::stepsToInt(temp_vec);
} else {
all_rows[0][submatrix_j] = initialVector;
}
}
// padding string a step vectors
for (int submatrix_i = 1; submatrix_i <= row_num; submatrix_i++) {
if ((submatrix_i * submatrix_dim - 1) >= string_a_real_size) {
vector<int> temp_vec(submatrix_dim, 0);
for (int i = 0;
i < (string_a_real_size - ((submatrix_i - 1) * submatrix_dim));
i++)
temp_vec[i] = 1;
all_columns[submatrix_i][0] =
SubmatrixCalculator::stepsToInt(temp_vec);
} else {
all_columns[submatrix_i][0] = initialVector;
}
}
for (int submatrix_i = 1; submatrix_i <= row_num; submatrix_i++) {
if ((submatrix_i - 1) * submatrix_dim > string_a_real_size)
top_left_costs[submatrix_i][1] = string_a_real_size;
else
top_left_costs[submatrix_i][1] = (submatrix_i - 1) * submatrix_dim;
for (int submatrix_j = 1; submatrix_j <= column_num; submatrix_j++) {
pair<int, int> final_steps = subm_calc->resultIndex[
// offset calculation
str_a_offsets[submatrix_i] + // left string
str_b_offsets[submatrix_j] + // top string
// left steps
subm_calc->stepOffsets[0][all_columns[submatrix_i][submatrix_j - 1]] +
// top steps
subm_calc->stepOffsets[1][all_rows[submatrix_i - 1][submatrix_j]]
];
all_columns[submatrix_i][submatrix_j] = final_steps.first;
all_rows[submatrix_i][submatrix_j] = final_steps.second;
if (submatrix_j != 1) {
top_left_costs[submatrix_i][submatrix_j] =
top_left_costs[submatrix_i][submatrix_j - 1];
top_left_costs[submatrix_i][submatrix_j] += subm_calc->sumSteps(
all_rows[submatrix_i - 1][submatrix_j - 1]);
}
}
}
}
Within the function itself there are many nested loops with calls to the sumSteps function in the SubmatrixCalculator.cpp file. Since the purpose of this function is to retrieve the path through the matrix while calculating the number of steps, this simple operation could benefit from the usage of a GPU rather than a CPU.
Recommendation
Based on the above profiling, our group has decided to go with Sudoku solver as our program to parallelize.
In order to determine the potential speed up for the Sudoku solver, we utilized Amdahl's Law:
S480 = 1 / ( 1 - 0.47 + 0.47 / 480 ) = 1.88
At best, we expect the process time to drop from 17.13 secs to about 17.13 / 1.88 = 9.11 secs
CheckSquare() O(n) = 8n^3 + 3n^2 + 10n CheckColumn() O(n) = 6n^2 + 4n CheckRow() O(n) = 6n^2 + 4n Print() O(n) = 4n^3 + 3n^2 + 3n StorePositions() O(n) = 4n^3 + 3n^2 + 3n GoBack() O(n) = 11n PlaceNum() O(n) = 7n^2 + 5n SolveSudoku() O(n) = 8n^3 + 3n^2 + 2n Main() O(n) = 15n^3 + 7n^2 + 15n Entire program O(n) = 42n^3 + 38n^2 + 59n
The Big-O of the Sudoku program is O(n^3), or cubic. This program would be idea for parallalizing.
The reason we chose the Sudoku solver was because we felt that the Sequence Alignment was too complex of an algorithm to work with, and the image processor seemed a bit too simple to parallelize. The sudoku solver seemed to be the right balance in terms of complexity as well as potential speed up. We are nearly cutting the time in half for a standard 9x9 puzzle, but as the puzzles get harder, through either an increase in the matrix size, or through an increase in blank numbers to be solved, we would expect even better performance through parallel programming than standard serial programming.
Assignment 2
Parallelized Oil Painting Program
Despite our initial decision of choosing to parallelize the Sudoku solver in assignment 1, we came to the conclusion that parallelizing the oil painting program would be better suited for us. We were able to grasp the logic behind the oil painting program whereas we had a lot of trouble working out the logic behind solving Sudoku puzzles.
The Code
Media: A2-Blastoise.zip This is a zip file that contains our full code and an executable version. To create your own project in visual studio with this code, you will need to download OpenCV. You will need the following property settings in your project: add the include path of OpenCV to VC++ Directories -> Include Directories and also add opencv_world320d.lib to Linker -> Input and add a post build command to copy OpenCV dll files into your project directory. (The last step can also be done by copying OpenCV bin files into debug directory of your project.)
In the serial program, the oil painting worked by going one pixel at a time in a double for loop.
for (int i=0; i < height; i++)
{
for (int j=0; j < width; j++)
{
//pixel processing
}
}
We were able to remove the need for this loop through the utilization of a kernel. In the main function, we created the following block and grid and called our oil painting kernel. The ceil function was used to calculate the grid dimensions, so that based on the block size the entire image will be included in the block.
const dim3 block(ntpb, ntpb); // ntpb was calculated based on device property (maxThreadsDim). const dim3 grid(ceil((float)width / block.x), ceil((float)height / block.y), 1); oilPaint << <grid, block >> >(gpu_src, gpu_dst, width, height);
In our kernel, through the use of the primitive types, we are able to determine the exact position of the pixel in the 2D array. Now instead of iterating through every pixel in the image, each thread in the block will adjust its own pixel intensity. The overall logic of the code stayed the same. We moved the double for loop into kernel and used i and j to locate the pixel.
//2D Index of current thread const int i = blockIdx.x * blockDim.x + threadIdx.x; const int j = blockIdx.y * blockDim.y + threadIdx.y;
The Results
The first graph is a comparison of the original execution time and the parallelize version. There was a considerable speed up. The original seems to have a growth rate that is exponential while the parallelized version is practically logarithmic. The second graph shows the time spent in the kernel in milliseconds, while also showing the percentage of time spend on the device verses host. The time spent in the kernel increases with the problem size, as expected. You can see that the time spent on the device also has a growth rate that is logarithmic, this means that for extremely small problem sizes the parallel version might not have a great speed up. (This is caused by the amount of CUDA API calls.)
Assignment 3
Media: OilPaint.zip This is our complete optimized solution, including the executable, code, and an image. The original parallelized solution took 3 arguments, the brush size, intensity level, the file name. Our default brush size was 5, and intensity level was 20, these values would be good for testing. Our optimized solution took only 1 argument, the file name. To run the parallel and optimized examples, the OpenCV files, specifically opencv_world320d.dll needs to be in the same directory as the executable. We tried multiple optimization method, some of them worked while others made our times worse. Below we have all our attempts at optimizations, what worked and what made it worse.
We attempted two potential applications of shared memory. Our first try was to store the entire image in shared memory. We realized that there was no need because this source image was only accessed once in the algorithm. Therefore transferring the image from global to shared, and then back to global, would be very inefficient. To make shared memory efficient, we would need to access it multiple times for any actual speedup.
Our second attempt was to store the array needed to calculate intensity. We realize that this would not be possible as each thread would require their own colour value arrays for calculations done at different times
Constant memory
We declared our image as constant memory. Constant memory was implemented and that increased our speed. However because the size of the image is dynamic, this can't be considered real constant memory.
Instruction mixing
For instruction mixing, we unrolled the inner loop with a constant brush size. We removed the option for the user to change the brush size and unrolled the loop with a constant brush size. We have so many checks and other functions inside of our inner loop that unrolling was causing it to slow down slightly. The result was a slight speed down and therefore we did not use instruction mixing. However we kept some functions that slightly sped the processing up.
Reduce CUDA API calls
We attempted to use only 1 array to hold both the source and destination. Because the destination image calculations depend on data from the source, we cannot change the source as we go.
Coalesced Access
Our last attempt at optimization was to try using coalesced access. We switched the x and y grid values so that all the values are adjacent to each other in memory. This resulted in a 1.3x speedup when combined with constant memory and occupancy optimization.
Optimization results
These are our results after different optimization steps. The combination of our optimizations resulted in a 1.3x speedup compared to our un-optimized version.